Abstract
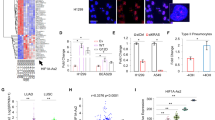
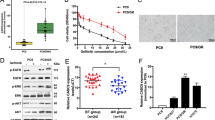
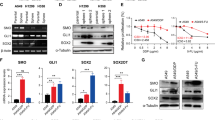
Recent advances in genomics unraveled several actionable mutational drivers in lung cancer, leading to promising therapies such as tyrosine kinase inhibitors and immune checkpoint inhibitors. However, the tumors’ acquired resistance to the newly-developed as well as existing therapies restricts life quality improvements. Therefore, we investigated the noncoding portion of the human transcriptome in search of alternative actionable targets. We identified an antisense transcript, LY6K-AS, with elevated expression in lung adenocarcinoma (LUAD) patients, and its higher expression in LUAD patients predicts poor survival outcomes. LY6K-AS abrogation interfered with the mitotic progression of lung cancer cells resulting in unfaithful chromosomal segregation. LY6K-AS interacts with and stabilizes 14-3-3 proteins to regulate the transcription of kinetochore and mitotic checkpoint proteins. We also show that LY6K-AS regulates the levels of histone H3 lysine 4 trimethylation (H3K4me3) at the promoters of kinetochore members. Cisplatin treatment and LY6K-AS silencing affect many common pathways enriched in cell cycle-related functions. LY6K-AS silencing affects the growth of xenografts derived from wildtype and cisplatin-resistant lung cancer cells. Collectively, these data indicate that LY6K-AS silencing is a promising therapeutic option for LUAD that inhibits oncogenic mitotic progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data associated with this publication have been deposited in GEO: GSE164419.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018;7:220–33.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60.
Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, Harel-Bellan A, et al. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5:e1257.
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311.
Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38.
Nowicki TS, Hu-Lieskovan S, Ribas A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J. 2018;24:47–53.
Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–23.
Kornienko AE, Dotter CP, Guenzl PM, Gisslinger H, Gisslinger B, Cleary C, et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016;17:14.
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208.
Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46.
Ohhata T, Senner CE, Hemberger M, Wutz A. Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev. 2011;25:1702–15.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63.
Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
Wu H, Yang L, Chen LL. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33:540–52.
Subhash S, Mishra K, Akhade VS, Kanduri M, Mondal T, Kanduri C. H3K4me2 and WDR5 enriched chromatin interacting long non-coding RNAs maintain transcriptionally competent chromatin at divergent transcriptional units. Nucleic Acids Res. 2018;46:9384–9400.
Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–79.
Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–37.
Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9.
Ali MM, Akhade VS, Kosalai ST, Subhash S, Statello L, Meryet-Figuiere M, et al. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun. 2018;9:883.
Marchese FP, Grossi E, Marin-Bejar O, Bharti SK, Raimondi I, Gonzalez J, et al. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell. 2016;63:397–407.
White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429.
Ooi AT, Gower AC, Zhang KX, Vick JL, Hong L, Nagao B, et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res. 2014;7:487–95.
Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967.
Isaka T, Nestor AL, Takada T, Allison DC. Chromosomal variations within aneuploid cancer lines. J Histochem Cytochem. 2003;51:1343–53.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Mahale S, Kumar M, Sharma A, Babu A, Ranjan S, Sachidanandan C, et al. The light intermediate chain 2 subpopulation of dynein regulates mitotic spindle orientation. Sci Rep. 2016;6:22.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Chen DY, Dai DF, Hua Y, Qi WQ. p53 suppresses 14-3-3gamma by stimulating proteasome-mediated 14-3-3gamma protein degradation. Int J Oncol. 2015;46:818–24.
Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–5.
Roeten MSF, Cloos J, Jansen G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother Pharmacol. 2018;81:227–43.
Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99.
Vedadi M, Blazer L, Eram MS, Barsyte-Lovejoy D, Arrowsmith CH, Hajian T. Targeting human SET1/MLL family of proteins. Protein Sci. 2017;26:662–76.
Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol. 2015;2:e970484.
Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–14.
Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–43.
Nishimura K, Johmura Y, Deguchi K, Jiang Z, Uchida KSK, Suzuki N, et al. Cdk1-mediated DIAPH1 phosphorylation maintains metaphase cortical tension and inactivates the spindle assembly checkpoint at anaphase. Nat Commun. 2019;10:981.
Lara-Gonzalez P, Moyle MW, Budrewicz J, Mendoza-Lopez J, Oegema K, Desai A. The G2-to-M transition is ensured by a dual mechanism that protects cyclin B from degradation by Cdc20-activated APC/C. Dev Cell. 2019;51:313–25.
Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW. Targeting mitosis in cancer: emerging strategies. Mol Cell. 2015;60:524–36.
Siemeister G, Mengel A, Fernandez-Montalvan AE, Bone W, Schroder J, Zitzmann-Kolbe S, et al. Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP Inhibitors In Vitro and In Vivo. Clin Cancer Res. 2019;25:1404–14.
Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–71.
Giannakakou P, Robey R, Fojo T, Blagosklonny MV. Low concentrations of paclitaxel induce cell type-dependent p53, p21 and G1/G2 arrest instead of mitotic arrest: molecular determinants of paclitaxel-induced cytotoxicity. Oncogene. 2001;20:3806–13.
Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22:688–95.
Hosing AS, Kundu ST, Dalal SN. 14-3-3 Gamma is required to enforce both the incomplete S phase and G2 DNA damage checkpoints. Cell Cycle. 2008;7:3171–9.
Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113:359–63.
Raungrut P, Wongkotsila A, Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, et al. Prognostic significance of 14-3-3gamma overexpression in advanced non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:3513–8.
Kasahara K, Goto H, Izawa I, Kiyono T, Watanabe N, Elowe S, et al. PI 3-kinase-dependent phosphorylation of Plk1-Ser99 promotes association with 14-3-3gamma and is required for metaphase-anaphase transition. Nat Commun. 2013;4:1882.
Bose A, Dalal SN. 14-3-3 proteins mediate the localization of Centrin2 to centrosome. J Biosci. 2019;44:42–10.
Kasahara K, Goto H, Enomoto M, Tomono Y, Kiyono T, Inagaki M. 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–12.
Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211.
Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–36.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Lassmann T, Hayashizaki Y, Daub CO. SAMStat: monitoring biases in next-generation sequencing data. Bioinformatics. 2011;27:130–1.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Subhash S, Kanduri C. GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. BMC Bioinfor. 2016;17:365.
Acknowledgements
This work was supported by the grants from Knut and Alice Wallenberg Foundation [KAW2014.0057]; Swedish Foundation for Strategic Research [RB13-0204]; Swedish Cancer Research foundation [Cancerfonden: Kontrakt no. CAN2018/591]; Swedish Research Council [2017-02834]; Barncancerfonden [PR2018-0090]; Ingabritt Och Arne Lundbergs forskningsstiftelse and LUA/ ALF (to CK). The Proteomics Core Facility at Gothenburg University, performed the analysis for protein identification. We acknowledge the Centre for Cellular Imaging at the University of Gothenburg for assisting in microscopy.
Author information
Authors and Affiliations
Contributions
Conceptualization—CK and MMA; Methodology—MMA, STK, SM, MDM, SR, DJ, CD, KM, and LS; Investigations—MMA, STK, SM, MDM, SR, DJ, CD, KM, and LS; Writing—original drafts- MMA; Writing—review and editing- MMA and CK; Funding acquisition, CK; Supervision, CK.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ali, M.M., Di Marco, M., Mahale, S. et al. LY6K-AS lncRNA is a lung adenocarcinoma prognostic biomarker and regulator of mitotic progression. Oncogene 40, 2463–2478 (2021). https://doi.org/10.1038/s41388-021-01696-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01696-7
This article is cited by
-
The pancancer overexpressed NFYC Antisense 1 controls cell cycle mitotic progression through in cis and in trans modes of action
Cell Death & Disease (2024)
-
LINC00460-FUS-MYC feedback loop drives breast cancer metastasis and doxorubicin resistance
Oncogene (2024)
-
H3K4me3-related lncRNAs signature and comprehensive analysis of H3K4me3 regulating tumor immunity in lung adenocarcinoma
Respiratory Research (2023)
-
The long non-coding RNA LINC00707 interacts with Smad proteins to regulate TGFβ signaling and cancer cell invasion
Cell Communication and Signaling (2023)
-
Gene amplification-driven lncRNA SNHG6 promotes tumorigenesis via epigenetically suppressing p27 expression and regulating cell cycle in non–small cell lung cancer
Cell Death Discovery (2022)