Abstract
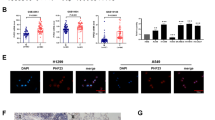
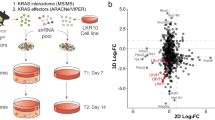
Radiation resistance is a major cause of lung cancer treatment failure. Armadillo (ARM) superfamily proteins participate in various fundamental cellular processes; however, whether ARM proteins regulate radiation resistance is not fully understood. Here, we used an unbiased CRISPR/Cas9 library screen and identified plakophilin 2 (PKP2), a member of the ARM superfamily of proteins, as a critical driver of radiation resistance in lung cancer. The PKP2 level was significantly higher after radiotherapy than before radiotherapy, and high PKP2 expression after radiotherapy predicted poor overall survival (OS) and postprogression survival (PPS). Mechanistically, mass spectrometry analysis identified that PKP2 was methylated at the arginine site and interacted with protein arginine methyltransferase 1 (PRMT1). Methylation of PKP2 by PRMT1 stabilized β-catenin by recruiting USP7, further inducing LIG4, a key DNA ligase in nonhomologous end-joining (NHEJ) repair. Concomitantly, PKP2-induced radioresistance depended on facilitating LIG4-mediated NHEJ repair in lung cancer. More strikingly, after exposure to irradiation, treatment with the PRMT1 inhibitor C-7280948 abolished PKP2-induced radioresistance, and C-7280948 is a potential radiosensitizer in lung cancer. In summary, our results demonstrate that targeting the PRMT1/PKP2/β-catenin/LIG4 pathway is an effective approach to overcome radiation resistance in lung cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rubin SM, Sage J. Manipulating the tumour-suppressor protein Rb in lung cancer reveals possible drug targets. Nature. 2019;569:343–4.
Nanavaty P, Alvarez MS, Alberts WM. Lung cancer screening: advantages, controversies, and applications. Cancer Control. 2014;21:9–14.
Kim BH, Kim YJ, Kim MH, Na YR, Jung D, Seok SH, et al. Identification of FES as a Novel Radiosensitizing Target in Human Cancers. Clin Cancer Res. 2020;26:265–73.
Coon D, Gokhale AS, Burton SA, Heron DE, Ozhasoglu C, Christie N. Fractionated stereotactic body radiation therapy in the treatment of primary, recurrent, and metastatic lung tumors: the role of positron emission tomography/computed tomography-based treatment planning. Clin Lung Cancer. 2008;9:217–21.
Chang JY, Liu YH, Zhu Z, Welsh JW, Gomez DR, Komaki R, et al. Stereotactic ablative radiotherapy: a potentially curable approach to early stage multiple primary lung cancer. Cancer. 2013;119:3402–10.
Jeong YK, Oh JY, Yoo JK, Lim SH, Kim EH. The Biofunctional Effects of Mesima as a Radiosensitizer for Hepatocellular Carcinoma. Int J Mol Sci. 2020;21:871.
Laird JH, Lok BH, Ma J, Bell A, de Stanchina E, Poirier JT, et al. Talazoparib Is a Potent Radiosensitizer in Small Cell Lung Cancer Cell Lines and Xenografts. Clin Cancer Res. 2018;24:5143–52.
Lee SY, Jeong EK, Ju MK, Jeon HM, Kim MY, Kim CH, et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16:10.
Gul IS, Hulpiau P, Saeys Y, van Roy F. Metazoan evolution of the armadillo repeat superfamily. Cell Mol Life Sci. 2017;74:525–41.
Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol. 1999;186:179–224.
Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–81.
Coates JC. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 2003;13:463–71.
Martin-Padron J, Boyero L, Rodriguez MI, Andrades A, Diaz-Cano I, Peinado P, et al. Plakophilin 1 enhances MYC translation, promoting squamous cell lung cancer. Oncogene. 2020;39:5479–93.
Li D, Song H, Mei H, Fang E, Wang X, Yang F, et al. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat Commun. 2018;9:2829.
Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–79.
Blanc RS, Richard S. Arginine Methylation: the Coming of Age. Mol Cell. 2017;65:8–24.
Suchankova J, Legartova S, Sehnalova P, Kozubek S, Valente S, Labella D, et al. PRMT1 arginine methyltransferase accumulates in cytoplasmic bodies that respond to selective inhibition and DNA damage. Eur J Histochem. 2014;58:2389.
Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–19.
Biau J, Chautard E, Verrelle P, Dutreix M, Altering DNA. Repair to Improve Radiation Therapy: specific and Multiple Pathway Targeting. Front Oncol. 2019;9:1009.
Jun S, Jung YS, Suh HN, Wang W, Kim MJ, Oh YS, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994.
Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta -catenin signaling. J Biol Chem. 2002;277:10512–22.
Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26.
Arimoto K, Burkart C, Yan M, Ran D, Weng S, Zhang DE. Plakophilin-2 promotes tumor development by enhancing ligand-dependent and -independent epidermal growth factor receptor dimerization and activation. Mol Cell Biol. 2014;34:3843–54.
Zhang B, Wu J, Cai Y, Luo M, Wang B, Gu Y. TCF7L1 indicates prognosis and promotes proliferation through activation of Keap1/NRF2 in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2019;51:375–85.
Wang Y, Chen X, Tang G, Liu D, Peng G, Ma W, et al. AS-IL6 promotes glioma cell invasion by inducing H3K27Ac enrichment at the IL6 promoter and activating IL6 transcription. FEBS Lett. 2016;590:4586–93.
Yu-Ju WuC, Chen CH, Lin CY, Feng LY, Lin YC, Wei KC, et al. CCL5 of glioma-associated microglia/macrophages regulates glioma migration and invasion via calcium-dependent matrix metalloproteinase 2. Neuro Oncol. 2020;22:253–66.
Zhou W, Wang K, Wang J, Qu J, Du G, Zhang Y. SOX17 Inhibits Tumor Metastasis Via Wnt Signaling In Endometrial Cancer. Onco Targets Ther. 2019;12:8275–86.
Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, et al. USP7 Is a Tumor-Specific WNT Activator for APC-Mutated Colorectal Cancer by Mediating beta-Catenin Deubiquitination. Cell Rep. 2017;21:612–27.
Su D, Ma S, Shan L, Wang Y, Wang Y, Cao C, et al. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J Clin Investig. 2018;128:4280–96.
Hsu JH, Hubbell-Engler B, Adelmant G, Huang J, Joyce CE, Vazquez F, et al. PRMT1-Mediated Translation Regulation Is a Crucial Vulnerability of Cancer. Cancer Res. 2017;77:4613–25.
Mertens C, Kuhn C, Franke WW. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J Cell Biol. 1996;135:1009–25.
Fischer-Keso R, Breuninger S, Hofmann S, Henn M, Rohrig T, Strobel P, et al. Plakophilins 1 and 3 bind to FXR1 and thereby influence the mRNA stability of desmosomal proteins. Mol Cell Biol. 2014;34:4244–56.
Zhang D, Qian Y, Liu X, Yu H, Zhao N, Wu Z. Up-regulation of plakophilin-2 is correlated with the progression of glioma. Neuropathology. 2017;37:207–16.
Niell N, Larriba MJ, Ferrer-Mayorga G, Sanchez-Perez I, Cantero R, Real FX, et al. The human PKP2/plakophilin-2 gene is induced by Wnt/beta-catenin in normal and colon cancer-associated fibroblasts. Int J Cancer. 2018;142:792–804.
Takahashi H, Nakatsuji H, Takahashi M, Avirmed S, Fukawa T, Takemura M, et al. Up-regulation of plakophilin-2 and Down-regulation of plakophilin-3 are correlated with invasiveness in bladder cancer. Urology. 2012;79:240 e241–248.
Demirag GG, Sullu Y, Gurgenyatagi D, Okumus NO, Yucel I. Expression of plakophilins (PKP1, PKP2, and PKP3) in gastric cancers. Diagn Pathol. 2011;6:1.
Hao XL, Tian Z, Han F, Chen JP, Gao LY, Liu JY. Plakophilin-2 accelerates cell proliferation and migration through activating EGFR signaling in lung adenocarcinoma. Pathol Res Pr. 2019;215:152438.
Vartak SV, Swarup HA, Gopalakrishnan V, Gopinatha VK, Ropars V, Nambiar M, et al. Autocyclized and oxidized forms of SCR7 induce cancer cell death by inhibiting nonhomologous DNA end joining in a Ligase IV dependent manner. FEBS J. 2018;285:3959–76.
Seol JH, Shim EY, Lee SE. Microhomology-mediated end joining: good, bad and ugly. Mutat Res. 2018;809:81–87.
Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst). 2014;17:110–20.
Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–60.
Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9.
Wu CT, Hsieh CC, Yen TC, Chen WC, Chen MF. TGF-beta1 mediates the radiation response of prostate cancer. J Mol Med (Berl). 2015;93:73–82.
Kim MR, Lee J, An YS, Jin YB, Park IC, Chung E, et al. TGFbeta1 protects cells from gamma-IR by enhancing the activity of the NHEJ repair pathway. Mol Cancer Res. 2015;13:319–29.
Kanamoto T, Hellman U, Heldin CH, Souchelnytskyi S. Functional proteomics of transforming growth factor-beta1-stimulated Mv1Lu epithelial cells: Rad51 as a target of TGFbeta1-dependent regulation of DNA repair. EMBO J. 2002;21:1219–30.
Dubash AD, Kam CY, Aguado BA, Patel DM, Delmar M, Shea LD, et al. Plakophilin-2 loss promotes TGF-beta1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes. J Cell Biol. 2016;212:425–38.
Adams MM, Wang B, Xia Z, Morales JC, Lu X, Donehower LA, et al. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle. 2005;4:1854–61.
Boisvert FM, Rhie A, Richard S, Doherty AJ. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle. 2005;4:1834–41.
Yang JH, Chiou YY, Fu SL, Shih IY, Weng TH, Lin WJ, et al. Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation. Nucleic Acids Res. 2014;42:9908–24.
Montenegro MF, Gonzalez-Guerrero R, Sanchez-del-Campo L, Pinero-Madrona A, Cabezas-Herrera J, Rodriguez-Lopez JN. Targeting the epigenetics of the DNA damage response in breast cancer. Cell Death Dis. 2016;7:e2180.
Montenegro MF, Gonzalez-Guerrero R, Sanchez-Del-Campo L, Pinero-Madrona A, Cabezas-Herrera J, Rodriguez-Lopez JN. PRMT1-dependent methylation of BRCA1 contributes to the epigenetic defense of breast cancer cells against ionizing radiation. Sci Rep.2020;10:13275.
Vadnais C, Chen R, Fraszczak J, Yu Z, Boulais J, Pinder J, et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat Commun. 2018;9:1418.
Acknowledgements
This study was supported by grants from the special fund for Jiangxi Key Laboratory (20171BCD40026 to YS), the Jiangxi Provincial Natural Science Foundation of China (20192BAB215039 to YS) and the Natural Science Foundation of China (81772821 to KH).
Author information
Authors and Affiliations
Contributions
CC, XP, SL, JY, CL and JT performed all of the experiments and data analysis. CC and YS conceived the research design, experiments, and data analysis. KH, GH, WM and YS prepared and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cheng, C., Pei, X., Li, SW. et al. CRISPR/Cas9 library screening uncovered methylated PKP2 as a critical driver of lung cancer radioresistance by stabilizing β-catenin. Oncogene 40, 2842–2857 (2021). https://doi.org/10.1038/s41388-021-01692-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01692-x
This article is cited by
-
PRMT1 in human neoplasm: cancer biology and potential therapeutic target
Cell Communication and Signaling (2024)
-
A review on CRISPR/Cas: a versatile tool for cancer screening, diagnosis, and clinic treatment
Functional & Integrative Genomics (2023)
-
CRISPR based therapeutics: a new paradigm in cancer precision medicine
Molecular Cancer (2022)
-
Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy
Molecular Cancer (2022)
-
PRMT1 and PRMT5: on the road of homologous recombination and non-homologous end joining
Genome Instability & Disease (2022)