Abstract
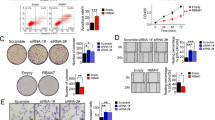
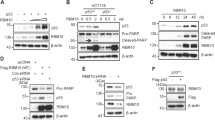
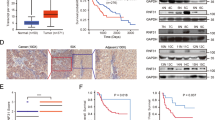
RNA-binding proteins play key roles in the posttranscriptional regulation of mRNA during cancer progression. Here, we show that RNA-binding motif protein 43 (RBM43) is significantly downregulated in human tumors, and its low expression is correlated with poor prognosis in patients with HCC. Overexpression of RBM43 suppressed cell proliferation in culture and resulted in the growth arrest of tumor xenografts, whereas downregulating RBM43 played an opposite role. We have also demonstrated that overexpression or knockdown of RBM43 affects the cell-cycle progression of liver cancer cells. Mechanistically, RBM43 directly associated with the 3′UTR of Cyclin B1 mRNA and regulated its expression. Moreover, loss of Rbm43 in mice promoted liver carcinogenesis and HCC development after diethylnitrosamine (DEN)-carbon tetrachloride (CCl4) treatment. Taken together, our data indicate that RBM43 is a tumor suppressor that controls the cell cycle through modulation of Cyclin B1 expression, providing evidence that RBM43 is particularly important in HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557–76.
Stefanie G, Markus H, Thomas T. A census of human RNA-binding proteins. Nat Rev Cancer. 2014;15:829–45.
Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327.
Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012;149:1393–406.
Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q, et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 2014;26:374–89.
Sakurai T, Isogaya K, Sakai S, Morikawa M, Morishita Y, Ehata S, et al. RNA-binding motif protein 47 inhibits Nrf2 activity to suppress tumor growth in lung adenocarcinoma. Oncogene 2016;35:5000.
Dang H, Takai A, Forgues M, Pomyen Y, Mou H, Xue W, et al. Oncogenic activation of the RNA binding protein NELFE and MYC signaling in hepatocellular carcinoma. Cancer Cell 2017;32:101–114.e108.
Zhao L, Cao J, Hu K, Wang P, Li G, He X, et al. RNA-binding protein RPS3 contributes to hepatocarcinogenesis by post-transcriptionally up-regulating SIRT1. Nucleic Acids Res. 2018;47:2011–28.
Guan M, Keaton JM, Dimitrov L, Hicks PJ, Xu J, Palmer ND, et al. Genome-wide association study identifies novel loci for type 2 diabetes-attributed end-stage kidney disease in African Americans. Hum Genom. 2019;13:21.
Albertsen HM, Chettier R, Farrington P, Ward K. Genome-wide association study link novel loci to endometriosis. PLoS ONE 2013;8:e58257.
Dong J, Levine DM, Buas MF, Zhang R, Onstad L, Fitzgerald RC, et al. Interactions between genetic variants and environmental factors affect risk of esophageal adenocarcinoma and Barrett’s esophagus. Clin Gastroenterol Hepatol. 2018;16:1598–1606.e1594.
Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72–75.
Dapito DH, Mencin A, Gwak G-Y, Pradere J-P, Jang M-K, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21:504–16.
Nurse P. Universal control mechanism regulating onset of M-phase. Nature 1990;344:503.
Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 2000;19:2340–50.
Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, et al. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120:865–80.
Takahashi K, Ishii Kana, Masakane Yamashita. Staufen1, Kinesin1 and microtubule function in cyclin B1 mRNA transport to the animal polar cytoplasm of zebrafish oocytes. Biochem Biophys Res Commu. 2018;503:2778–83.
Van Assche E, Van Puyvelde S, Vanderleyden J, Steenackers HP. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol. 2015;6:141.
Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–86.
Hartwell LH, Kastan MB. Cell cycle control and cancer. Science 1994;266:1821–8.
Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316.
Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature 2001;411:342.
Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18:851–5.
Aaltonen K, Amini RM, Heikkila P, Aittomaki K, Tamminen A, Nevanlinna H, et al. High cyclin B1 expression is associated with poor survival in breast cancer. Br J Cancer. 2009;100:1055–60.
Zhang H, Zhang X, Li X, Meng WB, Bai ZT, Rui SZ, et al. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J Cell Physiol. 2019;234:619–31.
EGLOFF AM, WEISSFELD J, SR LAND, FINN OJ. Evaluation of anticyclin B1 serum antibody as a diagnostic and prognostic biomarker for lung cancer. Ann NY Acad Sci. 2005;1062:29–40.
Covini G, Chan E, Nishioka M, Morshed SA, Reed SI, Tan EM. Immune response to cyclin B1 in hepatocellular carcinoma. Hepatology 1997;25:75–80.
Wang Z, Fan M, Candas D, Zhang TQ, Qin L, Eldridge A, et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell 2014;29:217–32.
Acknowledgements
We thank professor Ting Ni from the School of Life Sciences, Fudan University for advice in the analysis of RBM43 target genes. We thank Gang Wei, Ziya Re, and Yicheng Le (School of Life Sciences, Fudan University) for their helpful discussions and comments. We would like to thank Changjuan Shao, Yahui Zhao, Dan Song, Juanjuan Wang, Xiaoding Hu, Yi He, and Lifang You and Bingying Li (School of Life Sciences, Fudan University) for their advice and assistance in experimental skills and helpful discussions and comments.
Funding
This work was supported by the National Natural Science Foundation of China (81702742 to SS and 81972712 to JW).
Author information
Authors and Affiliations
Contributions
HF and SS designed the study, conducted most of the experiments and wrote the paper. JL, YQ, YL, XL, FZ, YW, YZ, CS, JW and DS provided advice about the experiments. WS, DJ and LY contributed to the paper completion. JW conceived the study, provided overall guidance, and contributed to the paper completion.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Feng, H., Liu, J., Qiu, Y. et al. RNA-binding motif protein 43 (RBM43) suppresses hepatocellular carcinoma progression through modulation of cyclin B1 expression. Oncogene 39, 5495–5506 (2020). https://doi.org/10.1038/s41388-020-1380-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1380-7
This article is cited by
-
CDCA8 induced by NF-YA promotes hepatocellular carcinoma progression by regulating the MEK/ERK pathway
Experimental Hematology & Oncology (2023)
-
Bioinformatics screening the novel and promising targets of curcumin in hepatocellular carcinoma chemotherapy and prognosis
BMC Complementary Medicine and Therapies (2022)
-
RAD6 Positively Affects Tumorigenesis of Esophageal Squamous Cell Carcinoma by Regulating Histone Ubiquitination of CCNB1
Biological Procedures Online (2022)
-
Elevated expression of the RNA‐binding motif protein 43 predicts poor prognosis in esophageal squamous cell carcinoma
International Journal of Clinical Oncology (2021)