Abstract
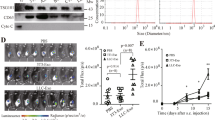
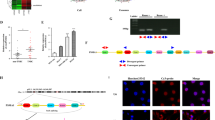
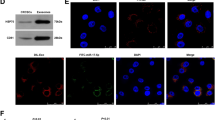
We previously identified that the development of early-stage myeloid-derived suppressor cells (eMDSCs) in breast cancer with high IL-6 (IL-6high) expression was correlated with the SOCS3 deficiency-dependent hyperactivation of the JAK/STAT signaling pathway. However, the regulatory mechanisms have not yet been elucidated. In this study, we aimed to investigate how the posttranscriptional regulation mediated by cancer exosome-derived miRNAs affected the JAK/STAT signaling pathway and the development of eMDSCs. Using miRNA microarray, we screened miR-9 and miR-181a which were exclusively upregulated in eMDSCs and inversely associated with SOCS3 expression. We found both miRNAs promoted the amplification of immature eMDSCs with the strong suppression on T-cell immunity in mice and humans. Furthermore, miR-9 and miR-181a promoted 4T1 tumor growth and immune escape via enhancing eMDSCs infiltration in situ. But miR-9 and miR-181a stimulated eMDSCs development by separately inhibiting SOCS3 and PIAS3, two crucial regulators in the negative feedback loop of the JAK/STAT signaling pathway. Elevated miR-9 and miR-181a in eMDSCs was derived from tumor-derived exosomes, and blocking the exosome release could fully attenuate the miRNA-mediated regulation on eMDSCs development. In summary, our findings indicated that tumor exosome-derived miR-9 and miR-181a activated the JAK/STAT signaling pathway via targeting SOCS3 and PIAS3, respectively, and thus promoted the expansion of eMDSCs which might provide potential therapeutic target for IL-6high breast cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pyzer AR, Cole L, Rosenblatt J, Avigan DE. Myeloid-derived suppressor cells as effectors of immune suppression in cancer. Int J Cancer. 2016;139:1915–26.
Zhao Y, Wu T, Shao S, Shi B, Zhao Y. Phenotype, development, and biological function of myeloid-derived suppressor cells. OncoImmunology. 2016;5:e1004983.
Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150.
Sade-Feldman M, Kanterman J, Klieger Y, Ish-Shalom E, Olga M, Saragovi A, et al. Clinical significance of circulating CD33+Cd11b+HLA-DR- myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clin Cancer Res. 2016;22:5661–72.
Jiang M, Chen J, Zhang W, Zhang R, Ye Y, Liu P, et al. Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells via suppression of suppressor of cytokine signaling 3 in breast cancer. Front Immunol. 2017;8:1840.
Zhang W, Jiang M, Chen J, Zhang R, Ye Y, Liu P, et al. SOCS3 suppression promoted the recruitment of CD11b(+)Gr-1(-)F4/80(-)MHCII(-) early-stage myeloid-derived suppressor cells and accelerated interleukin-6-related tumor invasion via affecting myeloid differentiation in breast cancer. Front Immunol. 2018;9:1699.
Pu S, Qin B, He H, Zhan J, Wu Q, Zhang X, et al. Identification of early myeloid progenitors as immunosuppressive cells. Sci Rep. 2016;6:23115.
Zhou Z, French DL, Ma G, Eisenstein S, Chen Y, Divino CM, et al. Development and function of myeloid-derived suppressor cells generated from mouse embryonic and hematopoietic stem cells. Stem Cells. 2010;28:620–32.
Yu J, Du W, Yan F, Wang Y, Li H, Cao S, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190:3783–97.
Yu J, Wang Y, Yan F, Zhang P, Li H, Zhao H, et al. Noncanonical NF-κB activation mediates STAT3-stimulated IDO upregulation in myeloid-derived suppressor cells in breast cancer. J Immunol. 2014;193:2574–86.
Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15:23.
Jiang M, Zhang WW, Liu P, Yu W, Liu T, Yu J. Dysregulation of SOCS-mediated negative feedback of cytokine signaling in carcinogenesis and its significance in cancer treatment. Front Immunol. 2017;8:70.
Boosani CS, Agrawal DK. Methylation and microRNA-mediated epigenetic regulation of SOCS3. Mol Biol Rep. 2015;42:853–72.
Isomoto H. Epigenetic alterations in cholangiocarcinoma-sustained IL-6/STAT3 signaling in cholangio- carcinoma due to SOCS3 epigenetic silencing. Digestion. 2009;79:2–8.
Tischoff I, Hengge UR, Vieth M, Ell C, Stolte M, Weber A, et al. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut. 2007;56:1047–53.
He B, You L, Uematsu K, Zang K, Xu Z, Lee AY, et al. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc Natl Acad Sci USA. 2003;100:14133–8.
Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2011;2:720–7.
Xu Z, Ji J, Xu J, Li D, Shi G, Liu F, et al. MiR-30a increases MDSC differentiation and immunosuppressive function by targeting SOCS3 in mice with B-cell lymphoma. FEBS J. 2017;284:2410–24.
Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Ther. 2019;27:518–30.
Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu X, et al. Interplay between microRNAs and the STAT3 signaling pathway in human cancers. Physiol Genomics. 2013;45:1206–14.
Zheng H-B, Zheng X-G, Liu B-P. miRNA-101 inhibits ovarian cancer cells proliferation and invasion by down-regulating expression of SOCS-2. Int J Clin Exp Med. 2015;8:20263–70.
Zhang X, Wang J, Cheng J, Ding S, Li M, Sun S, et al. An integrated analysis of SOCS1 down-regulation in HBV infection-related hepatocellular carcinoma. J Viral Hepat. 2014;21:264–71.
Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–53.
Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–303.
Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X, et al. MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J Immunol. 2015;195:1301–11.
McClure C, McPeak MB, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Stat3 and C/EBPbeta synergize to induce miR-21 and miR-181b expression during sepsis. Immunol Cell Biol. 2017;95:42–55.
Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–44.
Lao M, Shi M, Zou Y, Huang M, Ye Y, Qiu Q, et al. Protein inhibitor of activated STAT3 regulates migration, invasion, and activation of fibroblast-like synoviocytes in rheumatoid arthritis. J Immunol. 2016;196:596–606.
Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, et al. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J Biol Chem. 2003;278:21327–30.
Liu B, Gross M, ten Hoeve J, Shuai K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc Natl Acad Sci USA. 2001;98:3203–7.
Tussié-Luna MI, Bayarsaihan D, Seto E, Ruddle FH, Roy AL. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc Natl Acad Sci USA. 2002;99:12807–12.
Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D’Ippolito E, Cataldo A, et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312.
Yang L, Niu F, Yao H, Liao K, Chen X, Kook Y, et al. Exosomal miR-9 released from HIV Tat stimulated astrocytes mediates microglial migration. J Neuroimmune Pharm. 2018;13:330–44.
Bjørnetrø T, Redalen KR, Meltzer S, Thusyanthan NS, Samiappan R, Jegerschöld C, et al. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles. 2019;8:1567219.
Khatibi S, Babon J, Wagner J, Manton JH, Tan CW, Zhu H-J, et al. TGF-β and IL-6 family signalling crosstalk: an integrated model. Growth Factors. 2017;35:100–24.
Zhen J, Chen W, Zhao L, Zang X, Liu Y. A negative Smad2/miR-9/ANO1 regulatory loop is responsible for LPS-induced sepsis. Biomedicine Pharmacother. 2019;116:109016.
Bhatia A, Kumar Y. Cellular and molecular mechanisms in cancer immune escape: a comprehensive review. Expert Rev Clin Immunol. 2014;10:41–62.
Guo X, Qiu W, Liu Q, Qian M, Wang S, Zhang Z, et al. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene. 2018;37:4239–59.
Sporn JC, Katsuta E, Yan L, Takabe K. Expression of MicroRNA-9 is associated with overall survival in breast cancer patients. J Surg Res. 2019;233:426–35.
Orangi E, Motovali-Bashi M. Evaluation of miRNA-9 and miRNA-34a as potential biomarkers for diagnosis of breast cancer in Iranian women. Gene. 2019;687:272–9.
Jang MH, Kim HJ, Gwak JM, Chung YR, Park SY. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum Pathol. 2017;68:69–78.
Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao Z, et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene. 2016;35:1302–13.
Al-Mahmood S, Sapiezynski J, Garbuzenko OB, Minko T. Metastatic and triple-negative breast cancer: challenges and treatment options. Drug Deliv Transl Res. 2018;8:1483–507.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No.81872143, 81472473, 81772840), National Science and Technology Support Program of China (Grant No.2015BAI12B15), National Science and Technology Major Project (Grant No.2018ZX09201-015), Project of Science and Technology of Tianjin (Grant No.13ZCZCSY20300, 18JCQNJC82700) and Key Project of Tianjin Health and Family Planning Commission (Grant No.16KG126). We appreciate the efforts of Dr Juntian Liu and Dr Shixia Li of Department of Preventive Health Screening Center in sample collection. We thank Dr Xiubao Ren of Department of Biotherapy for support of Flow cytometry. We also thank Prof Weijia Zhang of Department of Medicine of Icahn School of Medicine at Mount Sinai for constructive comments to our manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, M., Zhang, W., Zhang, R. et al. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene 39, 4681–4694 (2020). https://doi.org/10.1038/s41388-020-1322-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1322-4
This article is cited by
-
Exploring the dynamic interplay between exosomes and the immune tumor microenvironment: implications for breast cancer progression and therapeutic strategies
Breast Cancer Research (2024)
-
Myeloid-derived suppressor cells in cancer: therapeutic targets to overcome tumor immune evasion
Experimental Hematology & Oncology (2024)
-
Tumorigenic and tumoricidal properties of exosomes in cancers; a forward look
Cell Communication and Signaling (2024)
-
Roles of exosomes in immunotherapy for solid cancers
Cell Death & Disease (2024)
-
Extracellular vesicle-based liquid biopsy biomarkers and their application in precision immuno-oncology
Biomarker Research (2023)