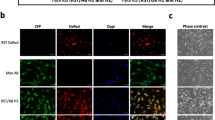
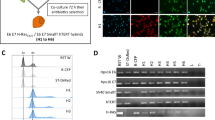
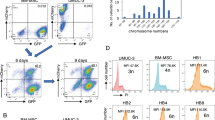
Abstract
Cell fusion in tumor progression mostly refers to the merging of a cancer cell with a cell that has migration and immune escape capabilities such as macrophages. Here we show that spontaneous hybrids made from the fusion of transformed mesenchymal cells with partners from the same lineage undergo nonrecurrent large-scale genomic rearrangements, leading to the creation of highly aneuploid cells with novel phenotypic traits, including metastatic spreading capabilities. Moreover, in contrast to their parents, hybrids were the only cells able to recapitulate in vivo all features of human pleomorphic sarcomas, a rare and genetically complex mesenchymal tumor. Hybrid tumors not only displayed specific mesenchymal markers, but also combined a complex genetic profile with a highly metastatic behavior, like their human counterparts. Finally, we provide evidence that patient-derived pleomorphic sarcoma cells are inclined to spontaneous cell fusion. The resulting hybrids also gain in aggressiveness, exhibiting superior growth capacity in mouse models. Altogether, these results indicate that cell fusion has the potential to promote cancer progression by increasing growth and/or metastatic capacities, regardless of the nature of the companion cell. Moreover, such events likely occur upon sarcoma development, paving the way for better understanding of the biology, and aggressiveness of these tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Massagué J, Batlle E, Gomis RR. Understanding the molecular mechanisms driving metastasis. Mol Oncol. 2017;11:3–4.
Riggi N, Aguet M, Stamenkovic I. Cancer metastasis: a reappraisal of its underlying mechanisms and their relevance to treatment. Annu Rev Pathol. 2018;13:117–40.
Pachmayr E, Treese C, Stein U. Underlying mechanisms for distant metastasis—molecular biology. Visc Med. 2017;33:11–20.
Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019. https://doi.org/10.1038/s41576-019-0114-6.
Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203.
Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58.
Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–62.
Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–7.
Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48:1119–30.
Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, et al. A renewed model of pancreas cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–82.
Turajlic S, Xu H, Litchfield K, Rowan A, Chambers T, Lopez JI, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173:581–94.e12.
Chibon F, Lagarde P, Salas S, Pérot G, Brouste V, Tirode F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–7.
Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Investig. 2004;113:913–23.
Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54.
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AAM, Voskuil DW et al. A gene-expression signature as a predictor of survival in breast cancer. http://dx.doi.org.gate2.inist.fr/10.1056/NEJMoa021967. 2009.
Delespaul L, Merle C, Lesluyes T, Lagarde P, Le Guellec S, Pérot G, et al. Fusion-mediated chromosomal instability promotes aneuploidy patterns that resemble human tumors. Oncogene. 2019. https://doi.org/10.1038/s41388-019-0859-6.
Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–8.
Rizvi AZ, Swain JR, Davies PS, Bailey AS, Decker AD, Willenbring H, et al. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci USA. 2006;103:6321–5.
Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, et al. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 2002;416:542–5.
Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901.
Weimann JM, Johansson CB, Trejo A, Blau HM. Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol. 2003;5:959–66.
Chakraborty A, Lazova R, Davies S, Bäckvall H, Ponten F, Brash D, et al. Donor DNA in a renal cell carcinoma metastasis from a bone marrow transplant recipient. Bone Marrow Transplant. 2004;34:183–6.
Gast CE, Silk AD, Zarour L, Riegler L, Burkhart JG, Gustafson KT, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4:eaat7828.
LaBerge GS, Duvall E, Grasmick Z, Haedicke K, Pawelek J. A melanoma lymph node metastasis with a donor-patient hybrid genome following bone marrow transplantation: a second case of leucocyte-tumor cell hybridization in cancer metastasis. PloS ONE. 2017;12:e0168581.
Lazova R, Laberge GS, Duvall E, Spoelstra N, Klump V, Sznol M, et al. A melanoma brain metastasis with a donor-patient hybrid genome following bone marrow transplantation: first evidence for fusion in human cancer. PloS ONE. 2013;8:e66731.
Duelli DM, Hearn S, Myers MP, Lazebnik Y. A primate virus generates transformed human cells by fusion. J Cell Biol. 2005;171:493–503.
Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol. 2007;17:431–7.
Mohr M, Zaenker KS, Dittmar T. Fusion in cancer: an explanatory model for aneuploidy, metastasis formation, and drug resistance. Methods Mol Biol. 2015;1313:21–40.
Zhou X, Merchak K, Lee W, Grande JP, Cascalho M, Platt JL. Cell fusion connects oncogenesis with tumor evolution. Am J Pathol. 2015;185:2049–60.
Broncy L, Paterlini-Bréchot P. Cancer-associated circulating atypical cells with both epithelial and macrophage-specific markers. J Lab Precis Med. 2018;3. http://jlpm.amegroups.com/article/view/4585. Accessed 18 Jun 2019.
Garvin S, Oda H, Arnesson L-G, Lindström A, Shabo I. Tumor cell expression of CD163 is associated to postoperative radiotherapy and poor prognosis in patients with breast cancer treated with breast-conserving surgery. J Cancer Res Clin Oncol. 2018;144:1253–63.
Laberge GS, Duvall E, Haedicke K, Pawelek J. Leukocyte–cancer cell fusion—genesis of a deadly journey. Cells. 2019;8. https://doi.org/10.3390/cells8020170.
Powell AE, Anderson EC, Davies PS, Silk AD, Pelz C, Impey S, et al. Fusion between Intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–505.
Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog. 2013;18:43–73.
Shabo I, Midtbö K, Andersson H, Åkerlund E, Olsson H, Wegman P, et al. Macrophage traits in cancer cells are induced by macrophage-cancer cell fusion and cannot be explained by cellular interaction. BMC Cancer. 2015;15:922.
Wang R, Sun X, Wang CY, Hu P, Chu C-Y, Liu S, et al. Spontaneous cancer-stromal cell fusion as a mechanism of prostate cancer androgen-independent progression. PloS ONE. 2012;7:e42653.
Xue J, Zhu Y, Sun Z, Ji R, Zhang X, Xu W, et al. Tumorigenic hybrids between mesenchymal stem cells and gastric cancer cells enhanced cancer proliferation, migration and stemness. BMC Cancer. 2015;15:793.
Lagarde P, Pérot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res J Am Assoc Cancer Res. 2012;18:826–38.
Lagarde P, Przybyl J, Brulard C, Pérot G, Pierron G, Delattre O, et al. Chromosome instability accounts for reverse metastatic outcomes of pediatric and adult synovial sarcomas. J Clin Oncol J Am Soc Clin Oncol. 2013;31:608–15.
Lartigue L, Neuville A, Lagarde P, Brulard C, Rutkowski P, Dei Tos P, et al. Genomic index predicts clinical outcome of intermediate-risk gastrointestinal stromal tumours, providing a new inclusion criterion for imatinib adjuvant therapy. Eur J Cancer. 2015;51:75–83.
Aichel O. Über Zellverschmelzung mit qualitativ abnormer Chromosomenverteilung als Ursache der Geschwulstbildung. W. Engelmann; 1911. https://books.google.com/books?id=8Sk4AQAAMAAJ.
Kerbel RS, Lagarde AE, Dennis JW, Donaghue TP. Spontaneous fusion in vivo between normal host and tumor cells: possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol Cell Biol. 1983;3:523–38.
Yilmaz Y, Lazova R, Qumsiyeh M, Cooper D, Pawelek J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005;35:1021–4.
Lazebnik Y. The shock of being united and symphiliosis. Another lesson from plants? Cell Cycle. 2014;13:2323–9.
Melzer C, von der Ohe J, Hass R. In vivo cell fusion between mesenchymal stroma/stem-like cells and breast cancer cells. Cancers. 2019;11. https://doi.org/10.3390/cancers11020185.
Searles SC, Santosa EK, Bui JD. Cell-cell fusion as a mechanism of DNA exchange in cancer. Oncotarget. 2018;9:6156–73.
Dodd RD, Mito JK, Kirsch DG. Animal models of soft-tissue sarcoma. Dis Model Mech. 2010;3:557–66.
Dodd RD, Añó L, Blum JM, Li Z, Van Mater D, Kirsch DG. Methods to generate genetically engineered mouse models of soft tissue sarcoma. Methods Mol Biol. 2015;1267:283–95.
Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–53.
Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–7.
Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–65.e28.
Chibon F, Mariani O, Derré J, Malinge S, Coindre JM, Guillou L, et al. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet. 2002;139:24–9.
Chibon F, Mariani O, Mairal A, Derré J, Coindre J-M, Terrier P, et al. The use of clustering software for the classification of comparative genomic hybridization data. an analysis of 109 malignant fibrous histiocytomas. Cancer Genet Cytogenet. 2003;141:75–8.
Gibault L, Pérot G, Chibon F, Bonnin S, Lagarde P, Terrier P, et al. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223:64–71.
Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch Int. J Pathol. 2010;456:201–17.
Mairal A, Terrier P, Chibon F, Sastre X, Lecesne A, Aurias A. Loss of chromosome 13 is the most frequent genomic imbalance in malignant fibrous histiocytomas. A comparative genomic hybridization analysis of a series of 30 cases. Cancer Genet Cytogenet. 1999;111:134–8.
Dittmar T, Zänker KS. Tissue regeneration in the chronically inflamed tumor environment: implications for cell fusion driven tumor progression and therapy resistant tumor hybrid cells. Int J Mol Sci. 2015;16:30362–81.
Berndt B, Zänker KS, Dittmar T. Cell fusion is a potent inducer of aneuploidy and drug resistance in tumor cell/ normal cell hybrids. Crit Rev Oncog. 2013;18:97–113.
Goldenberg DM. Horizontal transmission of malignancy by cell-cell fusion. Expert Opin Biol Ther. 2012;12 Suppl 1:S133–9.
Lu X, Kang Y. Cell fusion as a hidden force in tumor progression. Cancer Res. 2009;69:8536–9.
Dittmar T, Schwitalla S, Seidel J, Haverkampf S, Reith G, Meyer-Staeckling S, et al. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin Exp Metastasis. 2011;28:75–90.
Rachkovsky M, Sodi S, Chakraborty A, Avissar Y, Bolognia J, McNiff JM, et al. Melanoma x macrophage hybrids with enhanced metastatic potential. Clin Exp Metastasis. 1998;16:299–312.
Jemaà M, Abdallah S, Lledo G, Perrot G, Lesluyes T, Teyssier C, et al. Heterogeneity in sarcoma cell lines reveals enhanced motility of tetraploid versus diploid cells. Oncotarget. 2017;8:16669–89.
Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33:116–28.
Magdalou I, Lopez BS, Pasero P, Lambert SAE. The causes of replication stress and their consequences on genome stability and cell fate. Semin Cell Dev Biol. 2014;30:154–64.
Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–64.
Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–22.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37.
Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA. 2005;102:5992–7.
Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214.
Steele CD, Tarabichi M, Oukrif D, Webster AP, Ye H, Fittall M, et al. Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell. 2019;35:441–56.e8.
Han Z-Y, Richer W, Fréneaux P, Chauvin C, Lucchesi C, Guillemot D, et al. The occurrence of intracranial rhabdoid tumours in mice depends on temporal control of Smarcb1 inactivation. Nat Commun. 2016;7:10421.
Jiang E, Yan T, Xu Z, Shang Z. Tumor microenvironment and cell fusion. BioMed Res Int. 2019;2019:5013592.
Lagarde P, Brulard C, Pérot G, Mauduit O, Delespaul L, Neuville A, et al. Stable instability of sarcoma cell lines genome despite intra-tumoral heterogeneity: a genomic and transcriptomic study of sarcoma cell lines. Austin J Genet Genom Res. 2015;2:1014.
Durrieu-Gaillard S, Dumay-Odelot H, Boldina G, Tourasse NJ, Allard D, André F, et al. Regulation of RNA polymerase III transcription during transformation of human IMR90 fibroblasts with defined genetic elements. Cell Cycle. 2018;17:605–15.
Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, et al. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci USA. 2010;107:4176–81.
Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8.
Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–23.
Acknowledgements
This work was supported by the “Fondation ARC pour la recherche contre le cancer”, “the Fondation pour la recherche médicale” (FRM), and the associations “Phil’Anthrope” and “Pour Corentin”.
Author information
Authors and Affiliations
Contributions
LL, CM, and LD performed most of the experiments; PL generated hybrid and sarcoma cell lines; GP and LL performed some CGHs and immunostaining; TL performed the bioinformatics analyses; SLG and JMC did the histology analyses; LL and FC integrated and analyzed the data; LL and FC wrote the article with the help of CM; FC designed the study, supervised all the work, and obtained financial support.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lartigue, L., Merle, C., Lagarde, P. et al. Genome remodeling upon mesenchymal tumor cell fusion contributes to tumor progression and metastatic spread. Oncogene 39, 4198–4211 (2020). https://doi.org/10.1038/s41388-020-1276-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1276-6
This article is cited by
-
Single-cell landscape of undifferentiated pleomorphic sarcoma
Oncogene (2024)
-
Classical epithelial-mesenchymal transition (EMT) and alternative cell death process-driven blebbishield metastatic-witch (BMW) pathways to cancer metastasis
Signal Transduction and Targeted Therapy (2022)
-
Acquisition of cancer stem cell capacities after spontaneous cell fusion
BMC Cancer (2021)
-
Cell fusion enhances energy metabolism of mesenchymal tumor hybrid cells to sustain their proliferation and invasion
BMC Cancer (2021)
-
Cell–cell fusion of mesenchymal cells with distinct differentiations triggers genomic and transcriptomic remodelling toward tumour aggressiveness
Scientific Reports (2020)