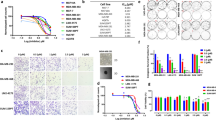
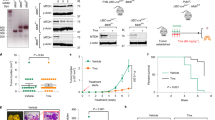
Abstract
SIRT5 is a member of the sirtuin family of NAD+-dependent protein lysine deacylases implicated in a variety of physiological processes. SIRT5 removes negatively charged malonyl, succinyl, and glutaryl groups from lysine residues and thereby regulates multiple enzymes involved in cellular metabolism and other biological processes. SIRT5 is overexpressed in human breast cancers and other malignancies, but little is known about the therapeutic potential of SIRT5 inhibition for treating cancer. Here we report that genetic SIRT5 disruption in breast cancer cell lines and mouse models caused increased succinylation of IDH2 and other metabolic enzymes, increased oxidative stress, and impaired transformation and tumorigenesis. We, therefore, developed potent, selective, and cell-permeable small-molecule SIRT5 inhibitors. SIRT5 inhibition suppressed the transformed properties of cultured breast cancer cells and significantly reduced mammary tumor growth in vivo, in both genetically engineered and xenotransplant mouse models. Considering that Sirt5 knockout mice are generally normal, with only mild phenotypes observed, these data establish SIRT5 as a promising target for treating breast cancer. The new SIRT5 inhibitors provide useful probes for future investigations of SIRT5 and an avenue for targeting SIRT5 as a therapeutic strategy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019;5:66.
Unterlass JE, Curtin NJ. Warburg and Krebs and related effects in cancer. Expert Rev Mol Med. 2019;21:e4.
Hirschey MD, Zhao Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol Cell Proteom: MCP. 2015;14:2308–15.
Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol. 2012;7:947–60.
Bheda P, Jing H, Wolberger C, Lin H. The substrate specificity of sirtuins. Annu Rev Biochem. 2016;85:405–29.
Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. https://doi.org/10.1038/nrm3293.
Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800.
Du J, Zhou Y, Su X, Yu J, Khan S, Jiang H, et al. Sirt5 is an NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–9.
Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell. 2015;59:321–32.
Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30.
Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteom: MCP. 2011;10:M111.012658.
Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–17.
Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1[alpha] destabilization. Cancer Cell. 2011;19:416–28.
Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11:253–70.
Sebastián C, Zwaans BMM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–99.
Chang L, Xi L, Liu Y, Liu R, Wu Z, Jian Z. SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol Med Rep. 2018;17:342–9.
Igci M, Kalender ME, Borazan E, Bozgeyik I, Bayraktar R, Bozgeyik E, et al. High-throughput screening of Sirtuin family of genes in breast cancer. Gene. 2016;586:123–8.
Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol: J Int Soc Oncodev Biol Med. 2014;35:10699–705.
Xu L, Che X, Wu Y, Song N, Shi S, Wang S, et al. SIRT5 as a biomarker for response to anthracycline-taxane-based neoadjuvant chemotherapy in triple-negative breast cancer. Oncol Rep. 2018;39:2315–23.
Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. 2018;9:545.
Xiangyun Y, Xiaomin N, Linping G, Yunhua X, Ziming L, Yongfeng Y, et al. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget. 2017;8:6984–93.
Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, et al. SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation. Cancer Res. 2018;78:372–86.
Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, et al. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191–5.
Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q, et al. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. 2016;17:811–22.
He B, Du J, Lin H. Thiosuccinyl peptides as Sirt5-specific inhibitors. J Am Chem Soc. 2012;134:1922–5.
Jiang Y, Zheng W. Cyclic tripeptide-based potent and selective human SIRT5 inhibitors. Med Chem. 2020;16:358–67.
Kalbas D, Liebscher S, Nowak T, Meleshin M, Pannek M, Popp C, et al. Potent and selective inhibitors of human Sirtuin 5. J Med Chem. 2018;61:2460–71.
Liu J, Huang Y, Zheng WA. Selective cyclic peptidic human SIRT5 inhibitor. Molecules. 2016;21:1217.
Maurer B, Rumpf T, Scharfe M, Stolfa DA, Schmitt ML, He W, et al. Inhibitors of the NAD+-dependent protein desuccinylase and demalonylase Sirt5. ACS Med Chem Lett. 2012;3:1050–3.
Rajabi N, Auth M, Troelsen KR, Pannek M, Bhatt DP, Fontenas M, et al. Mechanism-based inhibitors of the human Sirtuin 5 deacylase: structure-activity relationship, biostructural, and kinetic. Insight Angew Chem. 2017;56:14836–41.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–19.
Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76.
Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26.
Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–88.
Sadhukhan S, Liu X, Ryu D, Nelson OD, Stupinski JA, Li Z, et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc Natl Acad Sci USA. 2016;113:4320–5.
Greene KS, Lukey MJ, Wang X, Blank B, Druso JE, Lin MJ, et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proc Natl Acad Sci USA. 2019;116:26625–32.
Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–13.
Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8.
Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6.
Farooqi AS, Hong JY, Cao J, Lu X, Price IR, Zhao Q, et al. Novel lysine-based thioureas as mechanism-based inhibitors of Sirtuin 2 (SIRT2) with anticancer activity in a colorectal cancer murine model. J medicinal Chem. 2019;62:4131–41.
Spiegelman NA, Hong JY, Hu J, Jing H, Wang M, Price IR, et al. A small-molecule SIRT2 inhibitor that promotes K-Ras4a lysine fatty-acylation. ChemMedChem. 2019;14:744–8.
Jing H, Hu J, He B, Negrón Abril YL, Stupinski J, Weiser K, et al. A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell. 2016;29:297–310.
Smith BC, Denu JM. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry. 2007;46:14478–86.
Wagner GR, Bhatt DP, O’Connell TM, Thompson JW, Dubois LG, Backos DS, et al. A class of reactive Acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab. 2017;25:823–37. e828.
Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16.
Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–33.
Phan LM, Yeung SC, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11:1–19.
Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS. et al. Metabolic characterization of a Sirt5 deficient mouse model. Sci Rep. 2013;3:2806 https://doi.org/10.1038/srep02806.
Spiegelman NA, Price IR, Jing H, Wang M, Yang M, Cao J, et al. Direct comparison of SIRT2 inhibitors: potency, specificity, activity-dependent inhibition, and on-target anticancer activities. ChemMedChem. 2018;13:1890–4.
Acknowledgements
The authors thank the NCI Physical Sciences-Oncology Network Bioresource Core Facility (PBCF) for MDA-MB-231 cells, and Drs. Ruchika Bhawal and Sheng Zhang from the Cornell Proteomics and Metabolomics Facility for technical assistance. This work was supported in part by NIH R01 grants CA163255 and CA223534. YLNA was supported by NIH/NIGMS grant 5T32GM008500, and IRF was supported by NIH/NIGMS grant T32GM007273, a Cornell Deans Excellence Fellowship, and an HHMI Gilliam Fellowship. JA was supported by grants from the Ecole Polytechnique Federale de Lausanne (EPFL), and the Swiss National Science Foundation (SNSF 31003A_179435). This work utilized the Cornell University NMR facility, which is supported in part by the NSF through MRI award CHE-1531632.
Author information
Authors and Affiliations
Contributions
YLNA, IRF, JYH, RC, HL, and RSW conceptualization; YLNA, IRF, and JYH, formal analysis; YLNA, IRF, JYH, YLC, DAK, QZ, MY, JH, SS, BL, BH, BR, JJB, RD, JM, FW, VM, and TS investigation; YLNA, IRF, JYH, YLC, DAK, QZ, MY, JH, SS, BL, BH, and JJB methodology; YLNA, IRF, and JYH writing-original draft; YLNA, IRF, JYH, HL, and RSW writing-review and editing; JA, RC, HL, and RSW resources; RC, HL, and RSW funding acquisition; HL and RSW supervision; HL and RSW project administration.
Corresponding authors
Ethics declarations
Conflict of interest
Cornell University has patents on the SIRT5 inhibitors described in the manuscript. The authors have no additional competing interests to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Abril, Y.L.N., Fernandez, I.R., Hong, J.Y. et al. Pharmacological and genetic perturbation establish SIRT5 as a promising target in breast cancer. Oncogene 40, 1644–1658 (2021). https://doi.org/10.1038/s41388-020-01637-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01637-w
This article is cited by
-
Acylspermidines are conserved mitochondrial sirtuin-dependent metabolites
Nature Chemical Biology (2024)
-
A glimpse into novel acylations and their emerging role in regulating cancer metastasis
Cellular and Molecular Life Sciences (2024)
-
Oncometabolites drive tumorigenesis by enhancing protein acylation: from chromosomal remodelling to nonhistone modification
Journal of Experimental & Clinical Cancer Research (2022)