Abstract
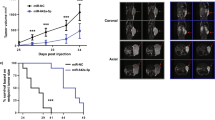
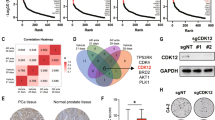
Available tools for prostate cancer (PC) prognosis are suboptimal but may be improved by better knowledge about genes driving tumor aggressiveness. Here, we identified FRMD6 (FERM domain-containing protein 6) as an aberrantly hypermethylated and significantly downregulated gene in PC. Low FRMD6 expression was associated with postoperative biochemical recurrence in two large PC patient cohorts. In overexpression and CRISPR/Cas9 knockout experiments in PC cell lines, FRMD6 inhibited viability, proliferation, cell cycle progression, colony formation, 3D spheroid growth, and tumor xenograft growth in mice. Transcriptomic, proteomic, and phospho-proteomic profiling revealed enrichment of Hippo/YAP and c-MYC signaling upon FRMD6 knockout. Connectivity Map analysis and drug repurposing experiments identified pyroxamide as a new potential therapy for FRMD6 deficient PC cells. Finally, we established orthotropic Frmd6 and Pten, or Pten only (control) knockout in the ROSA26 mouse prostate. After 12 weeks, Frmd6/Pten double knockouts presented high-grade prostatic intraepithelial neoplasia (HG-PIN) and hyperproliferation, while Pten single-knockouts developed only regular PIN lesions and displayed lower proliferation. In conclusion, FRMD6 was identified as a novel tumor suppressor gene and prognostic biomarker candidate in PC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30.
Steineck G, Helgesen F, Adolfsson J, Dickman PW, Johansson JE, Norlen BJ, et al. Quality of life after radical prostatectomy or watchful waiting. N. Engl J Med. 2002;347:790–6.
Gunn-Moore FJ, Tilston-Lunel AM, Reynolds PA. Willing to be involved in cancer. Genes. 2016;7:37. https://doi.org/10.3390/genes7070037.
Gunn-Moore FJ, Welsh GI, Herron LR, Brannigan F, Venkateswarlu K, Gillespie S, et al. A novel 4.1 ezrin radixin moesin (FERM)-containing protein, ‘Willin’. FEBS Lett. 2005;579:5089–94.
Ishiuchi T, Takeichi M. Nectins localize Willin to cell-cell junctions. Genes Cells. 2012;17:387–97.
Ishiuchi T, Takeichi M. Willin and Par3 cooperatively regulate epithelial apical constriction through aPKC-mediated ROCK phosphorylation. Nat Cell Biol. 2011;13:860–6.
Angus L, Moleirinho S, Herron L, Sinha A, Zhang X, Niestrata M, et al. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2012;31:238–50.
Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57.
Wang J, Hong Y, Shao S, Zhang K, Hong W. FFAR1-and FFAR4-dependent activation of Hippo pathway mediates DHA-induced apoptosis of androgen-independent prostate cancer cells. Biochem Biophys Res Commun. 2018;506:590–6.
Zhou PJ, Xue W, Peng J, Wang Y, Wei L, Yang Z, et al. Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway. J Exp Clin Cancer Res. 2017;36:139.
The Cancer Genome Atlas. https://www.cancer.gov/tcga.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43.
Long Q, Xu J, Osunkoya AO, Sannigrahi S, Johnson BA, Zhou W, et al. Global transcriptome analysis of formalin-fixed prostate cancer specimens identifies biomarkers of disease recurrence. Cancer Res. 2014;74:3228–37.
Haldrup C, Lynnerup AS, Storebjerg TM, Vang S, Wild P, Visakorpi T, et al. Large-scale evaluation of SLC18A2 in prostate cancer reveals diagnostic and prognostic biomarker potential at three molecular levels. Mol Oncol. 2016;10:825–37.
Haldrup C, Pedersen AL, Ogaard N, Strand SH, Hoyer S, Borre M et al. Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol Oncol. 2018;12:545–60.
Strand SH, Switnicki M, Moller M, Haldrup C, Storebjerg TM, Hedegaard J, et al. RHCG and TCAF1 promoter hypermethylation predicts biochemical recurrence in prostate cancer patients treated by radical prostatectomy. Oncotarget. 2017;8:5774–88.
Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat methods. 2014;11:599–600.
Zhu Y, Xu Y, Helseth DL, Jr. Gulukota K, Yang S, Pesce LL et al. Zodiac: a comprehensive depiction of genetic interactions in cancer by integrating TCGA data. J Natl Cancer Inst. 2015;107:djv129. https://doi.org/10.1093/jnci/djv129.
Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci USA. 2014;111:11139–44.
Correction for Pourdehnad et al. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proceedings of the National Academy of Sciences. 2013;110:17160.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52.e1417.
Moya IM, Halder G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol. 2019;20:211–26.
Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–27.
Riedel M, Berthelsen MF, Bakiri L, Wagner EF, Thomsen MK. Virus delivery of CRISPR guides to the murine prostate for gene alteration. J Vis Exp. 2018;57525. https://doi.org/10.3791/57525.
Dart DA, Uysal-Onganer P, Jiang WG. Prostate-specific PTen deletion in mice activates inflammatory microRNA expression pathways in the epithelium early in hyperplasia development. Oncogenesis. 2017;6:400.
Xu Y, Wang K, Yu Q. FRMD6 inhibits human glioblastoma growth and progression by negatively regulating activity of receptor tyrosine kinases. Oncotarget. 2016;7:70080–91.
Guan C, Chang Z, Gu X, Liu R. MTA2 promotes HCC progression through repressing FRMD6, a key upstream component of hippo signaling pathway. Biochem Biophys Res Commun. 2019;515:112–8.
Visser-Grieve S, Hao Y, Yang X. Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway. Oncogene. 2012;31:1189–95.
Bocci F, Tripathi SC, Vilchez Mercedes SA, George JT, Casabar JP, Wong PK, et al. NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr Biol. 2019;11:251–63.
Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–84.
Jolly MK, Somarelli JA, Sheth M, Biddle A, Tripathi SC, Armstrong AJ, et al. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharm Ther. 2019;194:161–84.
Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803.
Stauffer S, Chen X, Zhang L, Chen Y, Dong J. KIBRA promotes prostate cancer cell proliferation and motility. Febs J. 2016;283:1800–11.
Nishimoto M, Uranishi K, Asaka MN, Suzuki A, Mizuno Y, Hirasaki M, et al. Transformation of normal cells by aberrant activation of YAP via cMyc with TEAD. Sci Rep. 2019;9:10933.
Poole CJ, van Riggelen J. MYC-master regulator of the cancer epigenome and transcriptome. Genes. 2017;8:142. https://doi.org/10.3390/genes8050142.
Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–52.
Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J, et al. c-Myc modulation and acetylation is a key HDAC inhibitor target in cancer. Clin Cancer Res. 2017;23:2542–55.
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55.
Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15:222–34.
Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21.
Brawer MK. Prostatic intraepithelial neoplasia: an overview. Rev Urol. 2005;7:S11–18.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS ONE. 2014;9:e98187.
Feber A, Guilhamon P, Lechner M, Fenton T, Wilson GA, Thirlwell C, et al. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 2014;15:R30.
Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–30.
The R-project for statistical computing, 2014, https://www.r-project.org.
Schmidt L, Moller M, Haldrup C, Strand SH, Vang S, Hedegaard J et al. Exploring the transcriptome of hormone-naive multifocal prostate cancer and matched lymph node metastases. Br J Cancer. 2018;119:1527–37.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Harrell FE Jr., Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–6.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300.
Acknowledgements
Prof. Raymond C. Bergan (Department of Medicine, Northwestern University, Chicago, USA) generously provided the PC-3M cell line. The DU145-MN1 cell line was kindly provided by Dr Volker Jung (Institute of Urology and Pediatric Urology, University of Saarland, Hamburg/Saar, Germany). We thank Dr Søren Vang, Dr Morten Muhlig, and Dr Michael Knudsen for bioinformatics support. The Danish Cancer Biobank (DCB) is acknowledged for biological material. Ros Eeles and Zsofia Kote-Karai are supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden NHS Foundation Trust. Clara Cieza-Borella was supported by a donation from the Burstons and by research donations from the de Laszlo Foundation. This work was supported by The Danish Cancer Society (KDS), The Velux Foundation (KDS), The Novo Nordisk Foundation [NNF16OC0023048 (KDS); NNF16OC0022946 (MEJ); NNF14CC0001 (JVO)], The Independent Research Fund Denmark, The Harboe Foundation (JHJ), The Lundbeck Foundation [R231-2016-2682 (MEJ)], National Institute of Health Research (RE), The Biomedical Research Centre at the Institute of Cancer Research and Royal Marsden NHS Foundation Trust (RE), Richard and Debbi Burston, Nick Phillips and Bradshaw Foundation (CCB), and Aarhus University (JHJ/SHS).
Author information
Authors and Affiliations
Contributions
Conception and design: JHJ, SHS, and KDS. Development of methodology: JHJ, SHS, CCB, MT, and JVO. Acquisition of data JHJ, SHS, MR, SH, MN, MT, FDH, MB, and MEJ. Analysis and interpretation of data: JHJ, SHS, MEJ, MT, CCB, BU, and KDS. Writing of the manuscript: JHJ, SHS, and KDS. Revision and approval of the final manuscript: All authors. Study supervision: KDS, ZKJ, RE, and JVO.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haldrup, J., Strand, S.H., Cieza-Borrella, C. et al. FRMD6 has tumor suppressor functions in prostate cancer. Oncogene 40, 763–776 (2021). https://doi.org/10.1038/s41388-020-01548-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01548-w
This article is cited by
-
Exploring Prognostic Gene Factors in Breast Cancer via Machine Learning
Biochemical Genetics (2024)
-
ZEB1 hypermethylation is associated with better prognosis in patients with colon cancer
Clinical Epigenetics (2023)
-
FERM domain-containing protein FRMD6 activates the mTOR signaling pathway and promotes lung cancer progression
Frontiers of Medicine (2023)
-
CircRNA VPRBP inhibits tumorigenicity of cervical cancer via miR-93-5p/FRMD6 axis
Reproductive Sciences (2022)