Abstract
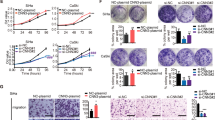
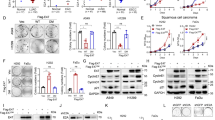
The molecular basis underlying the aggressive nature and excessive proliferation of cervical squamous cancer cell remains unclear. ΔNp63α is the predominant isotype of p63 expressed in the epithelia and regulates epithelial cell differentiation. The pro-/anti-tumor role of ΔNp63α in different kinds of solid tumors remains controversial and the precise molecular mechanisms are still elusive. In this study, we uncovered the molecular functions of ΔNp63α in cervical squamous cell carcinoma to clarify its roles as a tumor suppressor. We demonstrated that ΔNp63α suppressed cell migration, invasiveness, and tumor growth in SiHa and ME-180 cells with both in vivo and in vitro assays. Mechanistic investigation via RNA-sequencing and chromatin immunoprecipitation-sequencing revealed that ΔNp63α exerted its antitumor capacity via regulating the expression of a cohort of cell junction genes. Further, we showed that ZNF385B and CLDN1 were two direct ΔNp63α targets with significant relevance to cervical squamous cell carcinoma examined in cell cultures, tumor xenografts, and clinic tumors. We also demonstrated that ΔNp63α downregulated NFATC1 to reduce cisplatin resistance. These findings shed new lights on functions of ΔNp63α in tumors and providing novel insights in targeted therapy of cervical cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The accession number for the RNA-seq and ChIP-seq data reported in this paper is GEO: GSE135257.
References
Minion LE, Tewari KS. Cervical cancer—State of the science: from angiogenesis blockade to checkpoint inhibition. Gynecol Oncol. 2018;148:609–21.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Qian X, Ma C, Nie X, Lu J, Lenarz M, Kaufmann AM, et al. Biology and immunology of cancer stem(-like) cells in head and neck cancer. Crit Rev Oncol Hematol. 2015;95:337–45.
Rodríguez-Carunchio L, Soveral I, Steenbergen RD, Torné A, Martinez S, Fusté P, et al. HPV-negative carcinoma of the uterine cervix: a distinct type of cervical cancer with poor prognosis. BJOG. 2015;122:119–27.
Rusan M, Li YY, Hammerman PS. Genomic landscape of human papillomavirus-associated cancers. Clin Cancer Res. 2015;21:2009–19.
Rose PG, Bundy BN, Watkins EB, Thigpen JT, Deppe G, Maiman MA, et al. Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med. 1999;340:1144–53.
Sol ES, Lee TS, Koh SB, Oh HK, Ye GW, Choi YS. Comparison of concurrent chemoradiotherapy with cisplatin plus 5-fluorouracil versus cisplatin plus paclitaxel in patients with locally advanced cervical carcinoma. J Gynecol Oncol. 2009;20:28–34.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8.
Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. p63 in epithelial development. Cell Mol Life Sci. 2008;65:3126–33.
Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37:6092–104.
Ying H, Chang DL, Zheng H, McKeon F, Xiao ZX. DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol Cell Biol. 2005;25:6154–64.
Koster MI, Roop DR. p63 and epithelial appendage development. Differentiation. 2004;72:364–70.
Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–83.
Chen Y, Peng Y, Fan S, Li Y, Xiao ZX, Li C. A double dealing tale of p63: an oncogene or a tumor suppressor. Cell Mol Life Sci. 2018;75:965–73.
Flores ER. The roles of p63 in cancer. Cell Cycle. 2007;6:300–4.
Mills AA. p63: oncogene or tumor suppressor? Curr Opin Genet Dev. 2006;16:38–44.
Fisher ML, Kerr C, Adhikary G, Grun D, Xu W, Keillor JW, et al. Transglutaminase interaction with alpha6/beta4-Integrin stimulates YAP1-dependent ΔNp63α stabilization and leads to enhanced cancer stem cell survival and tumor formation. Cancer Res. 2016;76:7265–76.
Salah Z, Bar-mag T, Kohn Y, Pichiorri F, Palumbo T, Melino G, et al. Tumor suppressor WWOX binds to ΔNp63α and sensitizes cancer cells to chemotherapy. Cell Death Dis. 2013;4:e480.
Sen T, Sen N, Brait M, Begum S, Chatterjee A, Hoque MO, et al. ΔNp63α confers tumor cell resistance to cisplatin through the AKT1 transcriptional regulation. Cancer Res. 2011;71:1167–76.
Chung J, Lau J, Cheng LS, Grant RI, Robinson F, Ketela T, et al. SATB2 augments ΔNp63α in head and neck squamous cell carcinoma. EMBO Rep. 2010;11:777–83.
Dang TT, Esparza MA, Maine EA, Westcott JM, Pearson GW. ΔNp63α promotes breast cancer cell motility through the selective activation of components of the epithelial-to-mesenchymal transition program. Cancer Res. 2015;75:3925–35.
Ko E, Lee BB, Kim Y, Lee EJ, Cho EY, Han J, et al. Association of RASSF1A and p63 with poor recurrence-free survival in node-negative stage I-II non-small cell lung cancer. Clin Cancer Res. 2013;19:1204–12.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A mutant-p53/smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98.
Tran MN, Choi W, Wszolek MF, Navai N, Lee IL, Nitti G, et al. The p63 protein isoform ΔNp63α inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J Biol Chem. 2013;288:3275–88.
Dohn M, Zhang S, Chen X. p63alpha and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–205.
Zhou Y, Xu Q, Ling B, Xiao W, Liu P. Reduced expression of ΔNp63α in cervical squamous cell carcinoma. Clin Invest Med. 2011;34:E184–91.
Qian L, Xu F, Wang X, Jiang M, Wang J, Song W, et al. LncRNA expression profile of ΔNp63α in cervical squamous cancers and its suppressive effects on LIF expression. Cytokine. 2017;96:114–22.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8.
Si H, Lu H, Yang X, Mattox A, Jang M, Bian Y, et al. TNF-α modulates genome-wide redistribution of ΔNp63α/TAp73 and NF-κB c-REL interactive binding on TP53 and AP-1 motifs to promote an oncogenic gene program in squamous cancer. Oncogene. 2016;35:5781–94.
Rizzo JM, Oyelakin A, Min S, Smalley K, Bard J, Luo W, et al. Delta Np63 regulates IL-33 and IL-31 signaling in atopic dermatitis. Cell Death Differ. 2016;23:1073–85.
Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou HM, Rajagopal J, et al. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell. 2017;31:35–49.
Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006;580:4544–50.
Gerald RC, Eric NO. NFAT signaling. Cell. 2002;109:67–79.
Crotti TN, Flannery M, Walsh NC, Fleming JD, Goldring SR, McHugh KP. NFATc1 regulation of the human beta (3) integrin promoter in osteoclast differentiation. Gene. 2006;372:92–102.
Im JY, Lee KW, Won KJ, Kim BK, Ban HS, Yoon SH, et al. DNA damage-induced apoptosis suppressor (DDIAS), a novel target of NFATc1, is associated with cisplatin resistance in lung cancer. BBA-Mol Cell Res. 2016;1863:40–9.
Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27:6930–8.
Le Bras GF, Taubenslag KJ, Andl CD. The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adh Migr. 2012;6:365–73.
Martin TA, Jiang WG. Tight junctions and their role in cancer metastasis. Histol Histopathol. 2001;16:1183–95.
Szymborska A, Gerhardt H. Hold me, but not too tight—endothelial cell–cell junctions in angiogenesis. Cold Spring Harb Perspect Biol. 2018;10:a029223.
Hoevel T, Macek R, Mundigl O, Swisshelm K, Kubbies M. Expression and targeting of the tight junction protein CLDN1 in CLDN1-negative human breast tumor cells. J Cell Physiol. 2002;191:60–8.
Lv J, Sun BH, Mai ZT, Jiang MM, Du JF. CLDN-1 promoted the epithelial to migration and mesenchymal transition (EMT) in human bronchial epithelial cells via Notch pathway. Mol Cell Biochem. 2017;432:91–8.
Chao YC, Pan SH, Yang SC, Yu SL, Che TF, Lin CW, et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179:123–33.
Eftang LL, Esbensen Y, Tannaes TM, Blom GP, Bukholm IR, Bukholm G. Up-regulation of CLDN1 in gastric cancer is correlated with reduced survival. BMC Cancer. 2013;13:586.
Katayama A, Handa T, Komatsu K, Togo M, Horiguchi J, Nishiyama M, et al. Expression patterns of claudins in patients with triple-negative breast cancer are associated with nodal metastasis and worse outcome. Pathol Int. 2017;67:404–13.
Ratovitski EA. Phospho-Delta Np63 alpha regulates AQP3, ALOX12B, CASP14 and CLDN1 expression through transcription and microRNA modulation. FEBS Lett. 2013;587:3581–6.
Lopardo T, Lo Iacono N, Marinari B, Giustizieri ML, Cyr DG, Merlo G, et al. Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS ONE. 2008;3:e2715.
Lee M, Park J. Regulation of NFAT activation: a potential therapeutic target for immunosuppression. Mol Cells. 2006;22:1–7.
Pflaum J, Schlosser S, Muller M. p53 family and cellular stress responses in cancer. Front Oncol. 2014;4:285.
Machado-Silva A, Perrier S, Bourdon JC. p53 family members in cancer diagnosis and treatment. Semin Cancer Biol. 2010;20:57–62.
van Bokhoven H, Brunner HG. Splitting p63. Am J Hum Genet. 2002;71:1–13.
Qiao F, Bowie JU. The many faces of SAM. Sci's STKE. 2005;2005:re7.
Zhou Y, Wei Y, Zhu J, Wang Q, Bao L, Ma Y, et al. GRIM-19 disrupts E6/E6AP complex to rescue p53 and induce apoptosis in cervical cancers. PLoS ONE. 2011;6:e22065.
Chen L, Rashid F, Shah A, Awan HM, Wu MM, Liu A, et al. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. PNAS. 2015;112:10002–7.
Hu S, Wang X, Shan G. Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat Struct Mol Biol. 2016;23:1011–9.
Zhou Y, Li M, Wei Y, Feng D, Peng C, Weng H, et al. Down-regulation of GRIM-19 expression is associated with hyperactivation of STAT3-induced gene expression and tumor growth in human cervical cancers. J Interferon Cytokine Res. 2009;29:695–703.
National Research Council (US). Committee for the update of the guide for the care and use of laboratory animals. guide for the care and use of laboratory animals. 8th ed. Washington, DC: National Academies Press; 2011.
Acknowledgements
We would like to thank Yuhui Miao, Huimin Liu, Lu Qi, Xu Liu, Shuai Wei, Jingxin Li in University of Science and Technology of China for their valuable comments during the preparation of the manuscript. This work was supported by the National Natural Science Foundation of China (No. 81872110, 31600657, 81272881, 8137277"9, 81372777, 31725016, and 81902632); National Key Research and Development Program (2018YFC1003903); Anhui Provincial Key Research and Development Projects (1704a0802151); the Strategic Priority Research Program (Pilot study): “Biological basis of aging and therapeutic strategies” of the Chinese Academy of Sciences (grant XDPB10).
Author information
Authors and Affiliations
Contributions
Y.Z., G.S., X.L.W. and L.C. conceived the idea, designed the experiments, analyzed the data, wrote the paper with input from all authors and oversaw the project. F.X., L.L.Q. and J.W. performed the in vivo experiments. J.W., X.L.W. and F.X. performed the ChIP-seq, RNA-seq, qRT-PCR and analysis of the results. W.G.S. and D.Q.F. constructed the plasmids and established the stable cells. H.Y.L., Z.S., D.B.W. and B.L. performed IHC. H.Y.L. carried out most of the revision experiments. Y.Z., G.S. and L.C. wrote, reviewed, and/or revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance: ΔNp63α functions as a tumor suppressor in CSCC by regulating the expression of a cohort of cell junction genes and some other target genes such as CLDN1, ZNF385B, and NFATC1.
Rights and permissions
About this article
Cite this article
Zhou, Y., Liu, H., Wang, J. et al. ΔNp63α exerts antitumor functions in cervical squamous cell carcinoma. Oncogene 39, 905–921 (2020). https://doi.org/10.1038/s41388-019-1033-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-1033-x
This article is cited by
-
TP63–TRIM29 axis regulates enhancer methylation and chromosomal instability in prostate cancer
Epigenetics & Chromatin (2024)
-
∆Np63α inhibits Rac1 activation and cancer cell invasion through suppression of PREX1
Cell Death Discovery (2024)
-
SYT7 acts as an oncogene and a potential therapeutic target and was regulated by ΔNp63α in HNSCC
Cancer Cell International (2021)