Abstract
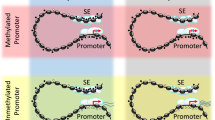
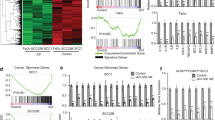
Head and neck squamous cell carcinoma (HNSCC) is a disease of significant morbidity and mortality and rarely diagnosed in early stages. Despite extensive genetic and genomic characterization, targeted therapeutics and diagnostic markers of HNSCC are lacking due to the inherent heterogeneity and complexity of the disease. Herein, we have generated the global histone mark based epigenomic and transcriptomic cartogram of SCC25, a representative cell type of mesenchymal HNSCC and its normal oral keratinocyte counterpart. Examination of genomic regions marked by differential chromatin states and associated with misregulated gene expression led us to identify SCC25 enriched regulatory sequences and transcription factors (TF) motifs. These findings were further strengthened by ATAC-seq based open chromatin and TF footprint analysis which unearthed Krüppel-like Factor 4 (KLF4) as a potential key regulator of the SCC25 cistrome. We reaffirm the results obtained from in silico and chromatin studies in SCC25 by ChIP-seq of KLF4 and identify ΔNp63 as a co-oncogenic driver of the cancer-specific gene expression milieu. Taken together, our results lead us to propose a model where elevated KLF4 levels sustains the oncogenic state of HNSCC by reactivating repressed chromatin domains at key downstream genes, often by targeting super-enhancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900.
Alorabi M, Shonka NA, Ganti AK. EGFR monoclonal antibodies in locally advanced head and neck squamous cell carcinoma: what is their current role? Crit Rev Oncol Hematol. 2016;99:170–9.
Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78.
Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60.
Iglesias-Bartolome R, Martin D, Gutkind JS. Exploiting the head and neck cancer oncogenome: widespread PI3K-mTOR pathway alterations and novel molecular targets. Cancer Discov. 2013;3:722–5.
Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–9.
Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13:4964–73.
Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3:770–81.
Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE. 2013;8:e56823.
Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–31.
Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8.
Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–55.
Li Q, Brown JB, Huang H, Bickel P. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 2011;5:1752–79.
Sethi I, Romano RA, Gluck C, Smalley K, Vojtesek B, Buck MJ, et al. A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genom. 2015;16:584.
Botchkarev VA, Flores ER. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb Perspect Med. 2014; 4.
Candi E, Amelio I, Agostini M, Melino G. MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 2015;22:12–21.
Sethi I, Sinha S, Buck MJ. Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genom. 2014;15:1042.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16.
Bishop JA, Teruya-Feldstein J, Westra WH, Pelosi G, Travis WD, Rekhtman N. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod Pathol. 2012;25:405–15.
Cui R, He J, Mei R, de Fromentel CC, Martel-Planche G, Taniere P, et al. Expression ofp53, p63, and p73 isoforms in squamous cell carcinoma and adenocarcinoma of esophagus. Biochem Biophys Res Commun. 2005;336:339–45.
He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11:73–78.
Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–90.
Sethi I, Gluck C, Zhou H, Buck MJ, Sinha S. Evolutionary re-wiring of p63 and the epigenomic regulatory landscape in keratinocytes and its potential implications on species-specific gene expression and phenotypes. Nucleic Acids Res. 2017;45:8208–24.
Jimenez L, Sharma VP, Condeelis J, Harris T, Ow TJ, Prystowsky MB, et al. MicroRNA-375 suppresses extracellular matrix degradation and invadopodial activity in head and neck squamous cell carcinoma. Arch Pathol Lab Med. 2015;139:1349–61.
Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol. 2011;27:185–211.
Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Nichols AC, Yoo J, Palma DA, Fung K, Franklin JH, Koropatnick J, et al. Frequent mutations in TP53 and CDKN2A found by next-generation sequencing of head and neck cancer cell lines. Arch Otolaryngol–head neck Surg. 2012;138:732–9.
Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22.
Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–60.
Evans PM, Liu C. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin. 2008;40:554–64.
Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, Grizzle W, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–34.
Jiang W, Lobo-Ruppert SM, Ruppert JM. Bad things happen in the basal layer: KLF4 and squamous cell carcinoma. Cancer Biol Ther. 2008;7:783–5.
Huang CC, Liu Z, Li X, Bailey SK, Nail CD, Foster KW, et al. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol Ther. 2005;4:1401–8.
Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500.
Tetreault MP, Wang ML, Yang Y, Travis J, Yu QC, Klein-Szanto AJ, et al. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology. 2010;139:2124–2134 e2129.
Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC, Chen CM. Nuclear Kruppel-like factor 4 expression is associated with human skin squamous cell carcinoma progression and metastasis. Cancer Biol Ther. 2008;7:777–82.
Tai SK, Yang MH, Chang SY, Chang YC, Li WY, Tsai TL, et al. Persistent Kruppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci. 2011;102:895–902.
Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2004;10:2709–19.
Kaichi S, Hasegawa K, Takaya T, Yokoo N, Mima T, Kawamura T, et al. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovasc Res. 2010;88:314–23.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72.
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11.
Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J Biol Chem. 2004;279:4862–8.
Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, et al. HOXB7 constitutively activates basic fibroblast growth factor in melanomas. Mol Cell Biol. 1996;16:4842–51.
Jones FS, Prediger EA, Bittner DA, De Robertis EM, Edelman GM. Cell adhesion molecules as targets for Hox genes: neural cell adhesion molecule promoter activity is modulated by cotransfection with Hox-2.5 and -2.4. Proc Natl Acad Sci USA. 1992;89:2086–90.
De Souza Setubal Destro MF, Bitu CC, Zecchin KG, Graner E, Lopes MA, Kowalski LP, et al. Overexpression of HOXB7 homeobox gene in oral cancer induces cellular proliferation and is associated with poor prognosis. Int J Oncol. 2010;36:141–9.
Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, et al. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. 2006;16:217–24.
Marcinkiewicz KM, Gudas LJ. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp Cell Res. 2014;320:128–43.
Marcinkiewicz KM, Gudas LJ. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J Cell Physiol. 2014;229:1405–16.
Yamatoji M, Kasamatsu A, Yamano Y, Sakuma K, Ogoshi K, Iyoda M, et al. State of homeobox A10 expression as a putative prognostic marker for oral squamous cell carcinoma. Oncol Rep. 2010;23:61–67.
Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–39.
Mao Z, Song S, Zhu Y, Yi X, Zhang H, Shang Y, et al. Transcriptional regulation of A33 antigen expression by gut-enriched Kruppel-like factor. Oncogene. 2003;22:4434–43.
Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–82.
Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–4002.
Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–90.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191.
Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Rahman M, Jackson LK, Johnson WE, Li DY, Bild AH, Piccolo SR. Alternative preprocessing of RNA-Sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics. 2015;31:3666–72.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Tomov ML, Olmsted ZT, Dogan H, Gongorurler E, Tsompana M, Otu HH, et al. Distinct and shared determinants of cardiomyocyte contractility in multi-lineage competent ethnically diverse human iPSCs. Sci Rep. 2016;6:37637.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Marinov GK, Kundaje A, Park PJ, Wold BJ. Large-scale quality analysis of published ChIP-seq data. G3. 2014;4:209–23.
Broad Institute. Picard. A set of command line tools (in Java) for manipulating high-throughput sequencing (HTS) data and formats such as SAM/BAM/CRAM and VCF. Broad Institute. http://broadinstitute.github.io/picard.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Bailey TL, Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128.
Lai WK, Bard JE, Buck MJ. ArchTEx: accurate extraction and visualization of next-generation sequence data. Bioinformatics. 2012;28:1021–3.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–7.
Thomas-Chollier M, Darbo E, Herrmann C, Defrance M, Thieffry D, van Helden J. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc. 2012;7:1551–68.
Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208.
Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–87.
Acknowledgements
We thank University at Buffalo Genomics and Bioinformatics Core facility for Next-generation sequencing and analysis support. These studies were supported, in part, by grants from the Community Foundation for Greater Buffalo to NJN.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tsompana, M., Gluck, C., Sethi, I. et al. Reactivation of super-enhancers by KLF4 in human Head and Neck Squamous Cell Carcinoma. Oncogene 39, 262–277 (2020). https://doi.org/10.1038/s41388-019-0990-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0990-4
This article is cited by
-
KLF7 regulates super-enhancer-driven IGF2BP2 overexpression to promote the progression of head and neck squamous cell carcinoma
Journal of Experimental & Clinical Cancer Research (2024)
-
The functions and prognostic value of Krüppel‐like factors in breast cancer
Cancer Cell International (2022)
-
ΔNp63 is a pioneer factor that binds inaccessible chromatin and elicits chromatin remodeling
Epigenetics & Chromatin (2021)