Abstract
At the molecular level, epithelial-to-mesenchymal transition (EMT) necessitates extensive transcriptional reprogramming which is orchestrated by a small group of gene-regulatory factors that include the zinc-finger DNA-binding protein SNAIL1. Although SNAIL1 is a well-known master regulator of EMT, knowledge of its immediate target genes is incomplete. Here, we used ChIP-seq to identify genes directly regulated by SNAIL1 in colorectal adenocarcinoma cells. When comparing the genomic distribution of SNAIL1 to that of the intestinal stem cell (ISC) transcription factors ASCL2 and TCF7L2, we observed a significant overlap. Furthermore, SNAIL1 ChIP-seq peaks are associated with a substantial fraction of ISC signature genes. In two colorectal cancer cell lines, we verified that SNAIL1 decreases ISC marker expression. Likewise, SNAIL1 directly represses the proto-oncogene MYB, and the long noncoding RNA (lncRNA) WiNTRLINC1, a recently described regulator of ASCL2. SNAIL1 targets multiple regulatory elements at the MYB and WiNTRLINC1 loci, and displaces ASCL2 and TCF7L2 from their binding regions at a MYB downstream regulatory element. Correlation analyses and expression profiling showed antiparallel expression of SNAIL1 and MYB in colorectal and breast cancer cell lines and tumor transcriptomes, suggesting that SNAIL1 controls MYB expression in different tissues. MYB loss-of-function attenuated proliferation and impaired clonogenicity in two- and three-dimensional cell cultures. Therefore, SNAIL1-mediated downregulation of MYB and ISC markers like WiNTRLINC1 likely contributes to the decrease in proliferation known to be associated with EMT, while simultaneously abrogating stemness features of colorectal cancer cells. Apparently, the relationship between EMT and stemness varies in different tumor entities.
Similar content being viewed by others
Introduction
Epithelial–mesenchymal transitions (EMT) are thought to facilitate several steps of the invasion–metastasis cascade [1,2,3]. Phenotypically, cells undergoing EMT lose their apical–basal polarity, tight intercellular contacts, and interactions with the basal membrane. They acquire a spindle-like appearance, gain motility, and invasiveness, and may acquire enhanced tumor-initiation capacities [1,2,3]. These radical changes in cellular traits demand for extensive transcriptional reprogramming, comprising down- and upregulation of epithelial and mesenchymal gene expression programs, respectively. Members of the SNAIL, ZEB and TWIST families of transcription factors are EMT master regulators which trigger these cascades in gene expression changes [1,2,3]. Although EMT-associated transcriptional adaptations are intensely investigated, their full extent and especially the complement of direct target genes of EMT master regulators have yet to be identified.
SNAIL1 proteins are evolutionary conserved zinc-finger transcription factors [4]. They recognize a specific DNA sequence motif 5′-CAGGTG-3′ which represents a variant of the E-box motif [5]. SNAIL1 proteins mainly act as transcriptional repressors, targeting, for instance, CDH1, coding for the cell–cell adhesion protein E-Cadherin, CLDN3, FOXA1, and the invasion suppressor EPHB3 [6,7,8,9]. Notably, SNAIL1 proteins can induce EMT in a variety of tissues, and different EMT programs may exist [10]. Therefore, an interesting question is whether transcriptional programs downstream of SNAIL1 are invariant or cell-type-specific. This could be addressed by comparing SNAIL1-bound cis-regulatory DNA elements and their associated genes in different cellular backgrounds. However, currently this information is available only for breast cancer (BRCA) EMT models [10,11,12].
EMT is commonly thought to enhance stemness of cancer cells [13,14,15,16], but contrasting results were also reported [9, 17, 18]. Further investigations aiming to clarify the relationship between stem cell properties and EMT are therefore needed. Colorectal cancer (CRC) appears well suited for this purpose. There is convincing evidence that the cells-of-origin in CRC are intestinal stem cells (ISCs) [19, 20]. ISCs have an epithelial character and are marked by the expression of a distinctive gene signature [21]. Furthermore, signal transduction pathways and transcription factors with crucial roles in ISC maintenance are known. Examples are the WNT/β-CATENIN pathway and its nuclear effector TCF7L2 [22, 23] which directly control the expression of many ISC signature genes [24,25,26,27,28]. This includes ASCL2 which codes for a basic helix-loop-helix transcription factor and plays an essential role in ISC fate decisions [22, 29]. Moreover, TCF7L2 and ASCL2 frequently co-occupy regulatory DNA elements at ISC signature genes and synergize in their regulation [29]. Altogether, the knowledge about ISC characteristics and their regulators provide excellent opportunities to examine the impact of EMT on stem cell features at a molecular level.
Here, we used chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) to determine the genomic distribution of murine SNAIL1 in colorectal adenocarcinoma cells with pronounced stem/progenitor character [9, 30]. SNAIL1 was found to occupy a significant number of ISC signature genes and to downregulate several of them. Specifically, the intestinal stemness-related genes WiNTRLINC1 and MYB are two newly identified genes which are directly repressed by SNAIL1. Furthermore, SNAIL1-bound regions frequently colocalize with sites occupied by TCF7L2 and ASCL2, and we provide evidence that SNAIL1 antagonizes TCF7L2 and ASCL2. Apparently, SNAIL1-induced EMT impairs stem cell features of CRC cells.
Results
Genome-wide mapping of Snail1-binding regions in CRC cells
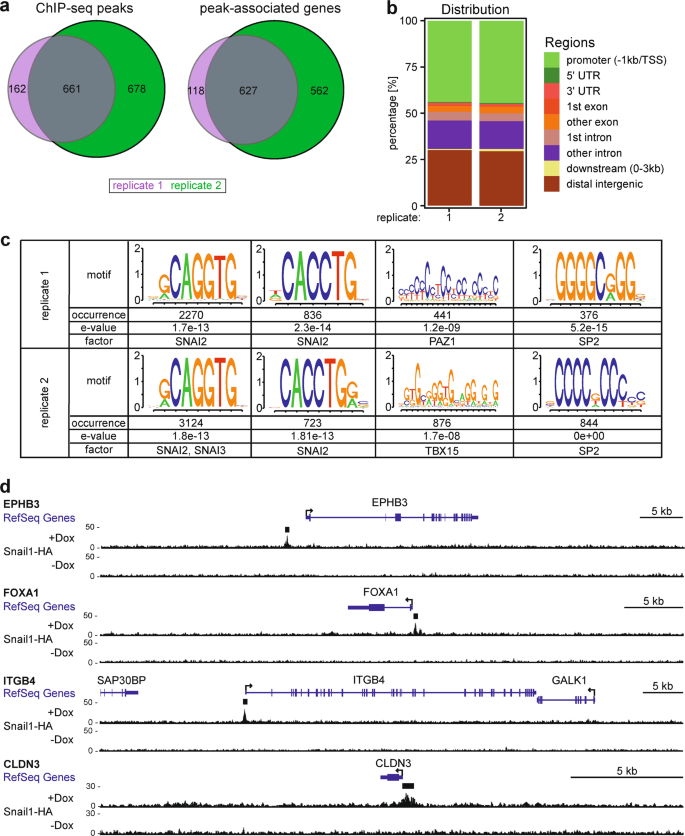
To identify genes that are directly regulated by SNAIL1 proteins, we expressed epitope-tagged murine SNAIL1 (Snail1-HA) in LS174T CRC cells from a doxycycline- (Dox-) inducible promoter [9], and performed ChIP-seq from untreated and Dox-treated cells. In two independent biological replicates, we mapped a total of 1501 Snail1-HA ChIP-seq peaks, 661 of which were identified in both replicates (Fig. 1a, Supplementary Table S1). Based on nearest neighbor relationships, Snail1-HA-bound regions were linked to 1307 genes, 627 of which were common to both replicates (Fig. 1a, Supplementary Table S1). Gene set enrichment analysis (GSEA) indicated that Snail1-HA ChIP-seq peak-associated genes were related to differentiation, morphogenesis, organogenesis, signaling, and cell junctions, which agrees well with the known biological functions of SNAIL1 proteins [1, 4] (Supplementary Fig. S1a, Supplementary Table S2).
Genome-wide identification of Snail1-HA DNA-binding sites in LS174T CRC cells. a Venn diagrams showing numbers of distinct and common ChIP-seq peaks and peak-associated genes from two independent experiments with LS174T cells stably transduced with retroviral expression vectors for Dox-inducible Snail1-HA. Cells were treated with 0.1 μg ml−1 Dox for 6 h prior to harvest and processing. b Distribution of Snail1-HA ChIP-seq peaks in replicates 1 and 2 across genomic regions. c Most abundant DNA sequence motifs identified in Snail1-HA ChIP-seq peaks from both replicates. Frequencies of motif occurrence, alignment quality (e-value), and transcription factors potentially recognizing the motifs are displayed. d Genome browser views of the indicated RefSeq gene loci depicting ChIP-seq tracks with called peaks (black bars) for Snail1-HA in LS174T cells in the presence or absence of Dox. Tracks represent a combination of replicates 1 and 2 and are based on hg19 sequence information
Overall, 44% of the Snail1-HA-bound regions were found within 1 kb upstream of transcriptional start sites (TSS). An additional 46% were located in intergenic regions and promoter-distal introns (Fig. 1b), indicating that Snail1-HA controls gene expression to a larger degree than commonly assumed from remote regulatory DNA elements. De novo motif deduction revealed the pervasive occurrence of three variants of the cognate SNAIL1 DNA-recognition motif within Snail1-HA ChIP-seq peaks [5]. 822/823 and 1332/1338 Snail1-HA ChIP-seq peaks in replicates 1 and 2, respectively, harbored such motifs considering an alignment score against the position weight matrix higher than 80% (Fig. 1c, Supplementary Fig. S1b). The only other DNA sequence elements enriched were highly G/C-rich (Fig. 1c) which may reflect the high G/C content of promoter regions (Fig. 1c). Genes associated with Snail1-HA ChIP-seq peaks included known SNAIL1 targets [7,8,9, 31], which are downregulated in Snail1-HA-expressing LS174T and HT29 cells (Fig. 1d, Supplementary Fig. S2) [8]. To investigate whether Snail1 regulates these genes when expressed at endogenous levels and to indirectly validate our mapping results in another cell model, we made use of MCF10A cells. These cells undergo SNAIL1-dependent EMT when treated with TGFβ1 [32]. In addition, SNAIL1 ChIP-on-chip data exist for MCF10A cells. We selected several genes, which were bound and regulated by Snail1-HA in LS174T cells, and whose promoter regions showed SNAIL1-occupancy also in MCF10A cells [12]. Indeed, concomitant with an increase in SNAIL1 expression, these genes are downregulated in TGFβ1-treated MCF10A cells (Supplementary Fig. 3), suggesting that SNAIL1 targets these genes not just when overexpressed. Based on these results the mapped ChIP-seq peaks appear to represent a high confidence collection of Snail1-HA binding regions.
To explore the regulatory impact of Snail1-HA occupancy we determined Snail1-HA-induced gene expression changes on a global scale and analyzed transcriptional responses of Snail1-HA-bound genes (Supplementary Fig. 4a, Supplementary Table S3). Overall, 8% of the genes associated with Snail1-HA ChIP-seq peaks were also significantly downregulated 6 h post Snail1-HA induction. The fraction of genes both deregulated and bound by Snail1-HA increased over time, and included a growing number of genes that were upregulated (Supplementary Fig. 4a, Supplementary Table S3). Overall, 44% of all Snail1-HA-binding events eventually translated into gene expression changes. These findings are in agreement with the notion that EMT is a process that gradually evolves [33], and that SNAIL1 proteins act mainly but not exclusively as transcriptional repressors [12, 34,35,36].
SNAIL1 proteins and their relative SLUG (SNAIL2) were implicated in EMT induction in different tissues and tumor entities. To explore to which extent Snail1-HA-binding regions and target genes are cell-type specific, and whether they are shared by SLUG, we compared the genome-wide distribution of Snail1 and Slug in human LS174T CRC cells and murine BRCA cells [10]. Even though we had identified some common targets in LS174T and MCF10A cells, the more comprehensive comparison based on ChIP-seq data showed that the number of Snail1-bound regions and their genic distribution differed considerably in the CRC and BRCA backgrounds (Supplementary Fig. S5, Supplementary Tables S4, S5). The binding pattern of Slug deviated even more, and showed a preponderance of promoter-distal introns and intergenic regions. Consistent with their divergent chromosomal distribution, only few genomic regions were bound by both Snail1 and Slug in human and mouse cells, respectively. We conclude that the chromosomal distribution patterns of Snail1 and Slug are largely cell-type- and factor-specific.
Snail1-HA ChIP-seq peaks colocalize with regions bound by ASCL2 and TCF7L2
To examine the relationship between Snail1-HA and ISC features we asked whether components of the murine ISC gene signature [21] were associated with Snail1-HA ChIP-seq peaks. Upon identifying the corresponding human genes it turned out that 11% of this signature had Snail1-HA ChIP-seq peaks in their vicinity (Fig. 2a, Supplementary Table S6). The regulatory impact of Snail1-HA on selected ISC signature genes was examined by quantitative reverse transcriptase PCR (qRT-PCR), demonstrating their downregulation by Snail1-HA (Supplementary Fig. S2). These results show that Snail1-HA interferes with the expression of stemness-associated genes in CRC cells.
Significant overlap of genomic-binding sites for Snail1-HA and the intestinal stem cell transcription factors ASCL2 and TCF7L2. a Overlap between genes associated with Snail1-HA ChIP-seq peaks in LS174T cells and components of the intestinal stem cell signature. b Census of regions exhibiting single occurrence, pairwise, and triple colocalization of Snail1-HA, ASCL2, and TCF7L2 ChIP-seq peaks as indicated by dots and vertical connector lines. For each factor, the total number of ChIP-seq peaks found in LS174T cells is shown in parentheses. Information for ASCL2 and TCF7L2 was derived from previously published data sets [29, 77]. Table on the right shows p-values for region-specific colocalization of Snail1-HA, ASCL2, and TCF7L2-binding events. c Genome browser views of RefSeq gene loci depicting the location of ChIP-seq peaks for Snail1-HA, ASCL2, and TCF7L2, and sequence conservation across 100 species (based on hg19 data). The central part shows a blowup of the region around 85 kb downstream of the MYB transcriptional start site (TSS). ChIP-seq peak regions for Snail1-HA, ASCL2, and TCF7L2 are marked by black bars. Regions where Snail1-HA, ASCL2, and TCF7L2 ChIP-seq peaks coincide, are highlighted by red framing
In addition, we compared the genome-wide binding patterns of Snail1-HA and the ISC factors ASCL2 and TCF7L2. We observed a significant colocalization of Snail1-HA, TCF7L2, and ASCL2 ChIP-seq peaks (Fig. 2b, Supplementary Table S7), including 274 regions upon which all three factors converge. Further comparison of patterns of transcription factor occupancy and gene expression changes revealed that 32% (1798/5662) and 31% (2067/6576) of all genes bound by ASCL2 and TCF7L2, respectively, were up- and downregulated in the presence of Snail1-HA (Supplementary Fig. S4b, Supplementary Table S3), most of them, however, apparently in an indirect manner. Interestingly, from a Snail1-HA point-of-view, between 51 and 61% of the genes bound by Snail1-HA and deregulated by Snail1-HA over a time course of 96 h showed co-occupancy by ASCL2. Among these, nearly half were additionally bound by TCF7L2 (Supplementary Fig. S4c, Supplementary Table S8). Only a minority of genes showed dual occupancy by Snail1-HA and TCF7L2. Moreover, even though nearly equal numbers of genes bound by ASCL2 and TCF7L2 were up- and downregulated in the presence of Snail1-HA (Supplementary Fig. S4b), those genes that were co-occupied by Snail1-HA, were predominantly repressed at every time point analyzed (Supplementary Fig. S4c).
The frequent colocalization of ChIP-seq peaks suggested that Snail1-HA might expel ASCL2 and TCF7L2 from their binding sites. To test this, we focused on two cases of coinciding ChIP-seq peaks at gene loci with functional importance for intestinal stemness (Fig. 2c). One is an intergenic element 85 kb downstream of the TSS of the MYB proto-oncogene. Despite its location, this element controls MYB expression in a TCF7L2-dependent manner for several reasons. First, deletion of the +85 kb element by using the CRISPR/Cas9 technology abrogates MYB but not AHI1 expression (Supplementary Fig. S6). Second, MYB, but not the adjacent AHI1 gene is repressed by Snail1-HA (Supplementary Fig. S7a), and third, knockout of TCF7L2 leads to a collapse of the active chromatin structure at the MYB +85 kb element and simultaneously reduces MYB but not AHI1 expression (Supplementary Fig. S7b, c). The second example is WiNTRLINC1 which codes for a long noncoding RNA controlling ASCL2 expression [37]. Expression analysis of WiNTRLINC1 and ASCL2 showed transient and long-lasting repression of both genes upon Snail1-HA induction in LS174T and HT29 cells, respectively (Supplementary Fig. S8b). Consistent with the regulatory interdependency between WiNTRLINC1 and ASCL2 [37] the two genes showed identical expression dynamics. We also verified binding of Snail1-HA at multiple positions around WiNTRLINC1 (Supplementary Fig. S8c) marking WiNTRLINC1 as novel Snail1-HA target. We then examined occupancy of selected ChIP-seq peaks at the WiNTRLINC1 locus and the MYB +85 kb element by Snail1-HA, ASCL2, and TCF7L2. Snail1-HA specifically associated with two ChIP-seq peaks at the WiNTRLINC1 locus, and the MYB +85 kb element, albeit levels of occupancy declined from 6 h to 24 h post induction (Supplementary Fig. S9c, d). In the absence of Snail1-HA, TCF7L2, and ASCL2 occupied all genomic regions examined. Upon Snail1-HA induction, ASCL2 and TCF7L2, which is not deregulated by Snail1-HA (Supplementary Fig. S10), dissociated from the MYB +85 kb element and disappeared almost completely over time. In contrast, at the WiNTRLINC1 locus, ASCL2 and TCF7L2 showed only a tendency to dissociation (Supplementary Fig. S9).
To find out whether Snail1-HA-induced dissociation of ASCL2 involves competition for the same binding sites, we performed electrophoretic mobility shift assays (EMSAs). At the WiNTRLINC1 locus, two of the three identified ASCL2-binding sites were indeed also recognized by Snail1-HA (Supplementary Fig. 11). At the MYB +85 kb region, however, Snail1-HA and ASCL2 interacted with nonidentical DNA elements (Supplementary Fig. 12). Sequence specificity of all interactions was demonstrated by mutagenesis of the 5′-CAGGTG-3′ and 5′-CAGCTG-3′ motifs, which resulted in loss of binding by Snail1-HA and ASCL2, respectively (Fig. 4c, Supplementary Figs. S11, S12). From these observations we conclude that Snail1-HA can displace ASCL2 from cis-regulatory DNA elements by competitive and noncompetitive mechanisms.
Inverse relationship of SNAI1 and MYB expression in colorectal and BRCA
Next, we focused on the regulatory relationship between SNAIL1 and MYB, and the contribution of MYB to stemness-related aspects of CRC cells. Pairwise-correlation analyses of gene expression data from CRC and BRCA samples revealed that MYB expression is positively correlated with that of epithelial marker genes (Fig. 3a, Supplementary Fig. S13a). In contrast, MYB expression is negatively correlated with that of EMT inducers and mesenchymal markers. Moreover, we found a significant survival advantage for patients with higher MYB levels when analyzing the combined TCGA colon and rectal adenocarcinoma cohorts and the pan-cancer cohort (Supplementary Fig. S13b). This, however, was not observed in BRCA samples (Supplementary Fig. S13b).
Expression of SNAIL1 and MYB is anticorrelated in colorectal and breast cancer. a Pairwise-correlation analyses of the expression of epithelial and mesenchymal marker genes in 443 colorectal tumor samples (GSE39582) and 466 breast tumor samples (TCGA). The red/blue color shading indicates the Pearson correlation coefficients as shown by the color bar. b, c qRT-PCR and western Blot analyses of MYB, SNAIL1, and SLUG expression in a cohort of CRC and two BRCA cell lines. Transcript levels are depicted as expression relative to GAPDH (rel. expr.). Shown are the mean and SEM; n = 3. GSK3β immunodetection was used as a loading control. MW molecular weight. d qRT-PCR analyses of Snail1-HA and MYB expression in LS174T and HT29 cells stably transduced with Dox-inducible retroviral control (vector) and Snail1-HA expression constructs. Cells were treated with 0.1 μg ml−1 Dox (LS174T) and 1 μg ml−1 Dox (HT29) for the indicated time periods. GAPDH was used for normalization and calculation of relative expression levels (rel. expr.) Shown are the mean and SEM; n = 3. e Western Blot analyses of Snail1-HA and MYB expression in LS174T and HT29 cells stably transduced with Dox-inducible retroviral control (vector) and Snail1-HA expression constructs. Cells were treated with 0.1 μg ml−1 Dox (LS174T) and 1 μg ml−1 Dox (HT29) for the indicated time periods. Detection of RNA polymerase II (POL II) and α-TUBULIN (TUBULIN) served as a loading control. MW: molecular weight
The inverse relationship between SNAI1 and MYB expression was verified in a panel of CRC cell lines and two BRCA cell lines with epithelial (MCF7) versus mesenchymal (MDA-MB-231) characteristics (Fig. 3b, c) [38]. Consistent with these anticorrelations, overexpression of Snail1-HA in CRC cells resulted in the rapid downregulation of MYB (Fig. 3d, e).
Expression of Snail1-HA reduces active chromatin features at the MYB locus
Aside from the MYB +85 kb element, ChIP-seq had identified two Snail1-HA-bound regions within MYB intron 1 (Figs. 2c and 4a). Association of Snail1-HA with these additional elements was confirmed by ChIP-qPCR (Fig. 4b) and EMSA (Fig. 4c, Supplementary Fig. S14). Thus, Snail1-HA can directly interact with specific DNA sequences at multiple positions of the MYB locus. To learn more about the mechanism whereby Snail1-HA represses MYB, we next investigated changes in chromatin structure and the abundance of histone marks typically found at active promoter and enhancer elements (H3K27ac), at poised and active enhancer elements (H3K4me1), and at promoter regions (H3K4me3). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) [39], we found an open chromatin conformation around the MYB TSS, and the +2.7 and +85 kb regions in control LS174T and HT29 cells (Fig. 4d). Upon expression of Snail1-HA, chromatin structure at these regions adopted a closed conformation in both cell lines.
MYB is a direct target gene of Snail1-HA. a Schematic representation of MYB exons 1 and 2, and a distal region 85 kb downstream of the transcriptional start site (TSS). The location of Snail1-HA ChIP-seq peaks, PCR amplicons employed in ChIP-qPCR and FAIRE analyses, as well as the positions of EMSA probes are indicated. Distance of PCR amplicons from the TSS is given in kilobase pairs (kb). b ChIP-qPCR experiments with LS174T and HT29 cells stably transduced with Dox-inducible retroviral control and Snail1-HA expression vectors. Cells were treated 0.1 µg ml−1 Dox (LS174T) and 1 µg ml−1 Dox (HT29) for 6 h. PCR amplicons as depicted in a. Shown are the mean and SEM; n = 3. c EMSA demonstrating binding of Snail1-HA to DNA sequences from MYB intron 1 in vitro. Material from in vitro translation reactions programmed with empty vector served as negative control (vector). WT wild type E-box motif, mut mutated E-box motif. d FAIRE analyses of MYB intron 1 and a region +85 kb downstream of the MYB gene in LS174T and HT29 cells stably transduced with Dox-inducible retroviral control and Snail1-HA expression vectors. Cells were left untreated or received 0.1 µg ml−1 Dox (LS174T) and 1 µg ml−1 Dox (HT29) for the times indicated. Data were calculated as relative enrichment of sequences of interest in formaldehyde-crosslinked versus non-crosslinked material. Shown are the mean and SEM; n = 3. e ChIP-qPCR analyses to assess the presence of H3K27Ac, H3K4me1, and H3K4me3 at different regions within intron 1 of the MYB gene and a region +85 kb downstream of the MYB TSS in LS174T and HT29 cells stably transduced with Dox-inducible retroviral control and Snail1-HA expression vectors. Cells were left untreated or received 0.1 µg ml−1 Dox (LS174T) and 1 µg ml−1 Dox (HT29) for the times indicated. Data were calculated as percent of input material. Enrichment was further normalized to histone H3 occupancy to account for regional differences in nucleosome density. Shown are the mean and SEM; n = 3. Statistical significance was calculated using a two-tailed unpaired Students’ t test. ***p-value < 0.001; **p-value < 0.01; *p-value < 0.05; ns: not significant
When we assessed histone marks at the MYB locus in the absence of Snail1-HA, we observed high levels of H3K27ac at several regions (Fig. 4e). Likewise, H3K4me3 was strongly represented around the TSS, whereas H3K4me1 accumulated at the MYB +2.7 and +85 kb regions, marking them as potential enhancer elements. Upon induction of Snail1-HA, H3K27ac levels diminished, especially at the +85 kb region. H3K4me1 and H3K4me3 seemingly were not affected by Snail1-HA. We conclude that Snail1-HA represses MYB by abolishing chromatin features characteristic for actively transcribed genes, but leaves the locus in a poised state.
Loss of MYB impairs viability and clonogenicity of CRC cells
Next, we investigated how knockdown and knockout of MYB affected the phenotype of two CRC cell lines (LS174T and LS411) with fairly high levels of MYB expression (Figs. 3 and 5; Supplementary Figs. S15, S16). MYB knockdown/knockout and control cells were analyzed with respect to viability, apoptosis, proliferation, and two- and three-dimensional colony formation. Since LS411 cells did not form colonies in soft agar, we assessed their three-dimensional growth using a limiting dilution assay with ultralow attachment plates. This additionally provides information about the frequency of sphere-forming units as measure of stem cell numbers [40]. MYB knockdown/knockout resulted in decreased MTT conversion (Fig. 5b, Supplementary Fig. S16b). This is likely due to impaired proliferation rather than increased cell death (Fig. 5c, d; Supplementary Fig. S16c, d). Furthermore, MYB knockdown/knockout diminished colony numbers in two-dimensional growth conditions (Fig. 5e; Supplementary Fig. S16e). Likewise, MYB loss-of-function impaired anchorage-independent growth (Fig. 5f; Supplementary Fig. S16f). Furthermore, stem cell frequency was significantly decreased. We did notice, though, that one of the three MYB knockout clones obtained, behaved like wild-type cells in all analyses. Possibly, 2F4 cells suffered additional genome changes that mask the consequences of MYB loss-of-function. Nonetheless, the functional analyses support an important role for MYB in the regulation of proliferation and clonogenicity of CRC cells.
Loss of MYB expression decreases viability, colony formation, and anchorage-independent growth of LS174T cells. a qRT-PCR and western Blot analyses of MYB expression in LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 96 h or were left untreated. Left: MYB transcript levels are depicted as expression relative to GAPDH (rel. expr.). Shown are the mean and SEM; n = 3. Right: Immunodetection of MYB. α-TUBULIN (TUBULIN) was used to control for equal loading. MW molecular weight. b The MTT assay with LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 96 h or were left untreated. Values of untreated cells were set to 1. Shown are the mean and SEM; n = 3. c Western Blot analyses of PARP1 and CASPASE3 cleavage in LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 96 h or were left untreated. Immunodetection of α-TUBULIN (TUBULIN) was used to control for equal loading. MW molecular weight. d Cell cycle analysis of LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 72 h or were left untreated. Cells were stained with propidium iodide (PI). Shown are the mean and SEM; n = 3. e The 2D colony formation assay of LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 12 days or were left untreated. Left: quantification of colony numbers after 12 days. Colonies were counted using ImageJ. Shown are the mean and SEM; n = 3. Right: representative image of a six-well plate with colonies stained with crystal violet after 12 days of incubation. f Anchorage-independent growth of LS174T cells stably transduced with lentiviral vectors for Dox-inducible shRNA expression. Cells were treated with 1 μg ml−1 Dox for 12 days or left untreated. Left: quantification of colony numbers using ImageJ. Shown are the mean and SEM; n = 3. Center: colony size. Diameters of at least 50 colonies from each condition were measured using ImageJ. Shown are the mean and SEM; n = 3 independent experiments. Right: representative images of colonies. Scale bar: 50 µm. a–f shNON non-silencing control shRNA, shMYB1, shMYB2 MYB-specific shRNAs. Statistical significance was calculated using a two-tailed unpaired Students’ t test. ***p-value < 0.001; **p-value < 0.01; *p-value < 0.05; ns: not significant
Discussion
Here, we determined the chromosomal distribution of Snail1-HA in a CRC EMT model to identify novel SNAIL1-regulated genes and to obtain deeper insights into gene-regulatory mechanisms and phenotypic changes associated with SNAIL1-induced EMT. Among the genes associated with Snail1-HA ChIP-seq peaks were several well-known SNAIL1 targets. This validated the reliability of our data set and allowed the confident identification of novel target genes such as WiNTRLINC1 and MYB. Furthermore, almost all Snail1-HA ChIP-seq peaks harbored the cognate SNAIL1 DNA-binding motif. In several cases we confirmed specific interactions of Snail1-HA with this motif in vitro. Apparently, in our model Snail1-HA interacts with the genome predominantly through its intrinsic sequence-specific DNA-binding capacity. In contrast, a recent study suggested that SNAIL1 may function through an alternative DNA sequence motif in BRCA cells [11]. This discrepancy could reflect direct versus indirect ways of chromosomal association. Such a piggyback mode of target gene access was previously reported for EMT inducers, possibly distinguishing repressive from activating target gene interactions [41, 42].
Comparison with previous studies indicates that SNAIL1 proteins occupy different genomic locations in different cellular backgrounds [10,11,12, 43, 44]. SNAIL1 and SLUG chromosomal distribution also differs despite identical DNA-binding specificity [5, 10]. Evidently, the range of DNA sequences that are occupied by SNAIL1 proteins and other EMT inducers is highly context-dependent. Contextual patterns of DNA occupancy could be determined by cell-type-specific interaction partners, but also by epigenetic factors. Significantly, regulatory elements bound by SNAIL1 proteins reside in an open chromatin conformation and are associated with active histone marks before the appearance of SNAIL1 proteins (this study, refs. [9, 12]). Accordingly, which genes and DNA elements are available for occupancy by SNAIL1 could be decisively determined by cell-type-specific preexisting chromatin landscapes. Likewise, Snail1-HA-induced gene repression was accompanied by changes in chromatin structural features (this study, refs. [9, 12]). Interestingly, at the MYB locus, H3K4me1, and H3K4me3 were unaffected by Snail1-HA. Apparently, MYB regulatory elements maintain a poised state during EMT which might allow for rapid MYB reactivation. This could be of significance for metastatic colonization which is thought to require mesenchymal-to-epithelial transition and resuscitation of epithelial gene expression [3].
Snail1-HA and ASCL2 ChIP-seq peaks frequently colocalized and both factors share highly similar DNA-recognition motifs [5]. Accordingly, Snail1-HA may displace ASCL2 and similar basic helix-loop-helix factors from regulatory DNA elements by competing for shared binding sites [9, 43]. This might also apply to the WiNTRLINC1 locus, even though expulsion of ASCL2 was incomplete at this gene. However, WiNTRLINC1 experienced only transient downregulation ins LS174T cells. Hence, the limited resolution of ChIP-qPCR experiments may have prevented detection of temporary ASCL2 dissociation from the WiNTRLINC1 locus. Thus, mutually exclusive occupancy of transcription factor binding sites likely represents an important mechanism whereby SNAIL1 proteins inactivate regulatory regions. The case of the MYB +85 kb element with nonoverlapping-binding sites for Snail1-HA and ASCL2 hints that SNAIL1 proteins engage additional, more indirect mechanisms for target inactivation, possibly based on chromatin structural changes [45, 46].
MYB, while best known for its role in hematopoiesis and hematological disorders, is also important for intestinal development, and functions as oncogene in solid cancers [47,48,49,50,51,52]. In agreement with its tumor-promoting capacity, MYB loss-of-function impaired viability, clonogenicity, and anchorage-independent growth of CRC cells. Nonetheless, quite counterintuitively, higher MYB expression levels correlated with a better prognosis for colorectal adenocarcinoma patients. Likewise, repression of MYB by Snail1-HA may seem surprising as this would attenuate oncogenic transformation. However, EMT is known to entail reduced cell proliferation [53] which most likely is a necessary corollary of increased motility. Thus, repression of MYB and possibly other oncogenes during EMT is plausible and could occur more frequently. Support for this comes from the observed anticorrelated expression of SNAIL1 and MYB in CRC and BRCA transcriptomes, and the repression of MYB also by ZEB1 [54]. Furthermore, comparatively higher MYB expression levels appear to be a characteristic of more epithelial tumors, while relatively lower MYB expression is found in more mesenchymal tumors, which in fact are those with a worse prognosis [55].
Knowledge about the cellular origin of CRCs and the extensive characterization of ISCs concerning essential transcription factors, key regulatory circuits of ISC fate, and distinguishing gene expression signatures [19,20,21,22,23, 56, 57] allowed us to examine at a molecular level how SNAIL1 proteins affect stemness features. The observed repression of multiple genes of crucial importance for ISCs [37, 56, 58] argues that SNAIL1-induced EMT interferes with stemness aspects of CRC cells. This is consistent with several reports demonstrating that stemness and tumor-initiating capacities are not necessarily linked to mesenchymal cell fates [9, 17, 18, 59]. Yet, SNAIL1 proteins and EMT have repeatedly been reported to promote stemness features of cancer cells [3, 13,14,15,16, 60, 61]. These contradictions might be reconciled in several ways. There might be different types of stemness installed by different combinations of transcription factors [10, 62]. Alternatively, a single genetic program that confers the defining properties of stem cells may be variably controlled by epithelial as well as mesenchymal collectives of transcription factors. Lastly, experimentally induced complete EMT may indeed abrogate stemness, whereas carcinogenesis possibly selects for more plastic, intermediate EMT states that amalgamate stemness aspects of epithelial cell states and higher mobility and invasiveness of mesenchymal cells [3].
In summary, our high confidence collection of SNAIL1-bound chromosomal regions represents a valuable resource for future studies aiming at a molecular-mechanistic dissection of EMT processes. As a paradigm, we analyzed the impact of SNAIL1 on a genetic program that underlies the ISC state. Our results hint at a complex interplay between EMT and stemness which warrants further investigations of the corresponding gene-regulatory circuits in cells with varying degrees of epithelial and mesenchymal characteristics.
Materials and methods
Cell culture
Cell lines (listed in Supplementary Table S9) were cultivated in DMEM with 10% (v/v) FCS, 10 mM HEPES, 1% (v/v) MEM nonessential amino acids, and 1% (v/v) penicillin/streptomycin at 37 °C and 5% CO2. MCF10A cells were cultivated in Advanced DMEM/F12 with 5% (v/v) Horse serum, 1% (v/v) (penicillin/streptomycin), 20 ng/ml human EGF, 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, and 10 μg/ml insulin at 37 °C and 5% CO2.
Oligonucleotides and antibodies
All oligonucleotides and antibodies used are listed in Supplementary Tables S10 and S11.
RNA isolation, cDNA synthesis, and qRT-PCR
RNA was isolated and reverse transcribed for quantitative gene expression by qRT-PCR as described [8] using GAPDH transcripts for normalization.
Protein extraction and western blotting
For detection of MYB, SNAIL1, and SLUG, nuclear extracts were prepared [8]. All other proteins were analyzed using whole cell lysates [28]. Detection was performed as described [63].
Dox-inducible shRNA expression
ShRNAs were selected based on top hits for MYB [64]. For cloning, 97mer oligonucleotides were PCR amplified, digested with EcoRI and XhoI and inserted into the pTRIPZ-vector (OpenBiosystems). The resulting lentiviral vectors were used for infection of LS174T cells as described [8]. Transduced cells were selected using 6 µg/ml blasticidin.
Genome editing
An online tool (http://crispr.mit.edu) [65] was employed to design gRNAs which were cloned into the gRNA expression vector (a gift from George Church, #41824, addgene, Cambridge, MA, USA) [66]. For genome editing, 2 × 106 cells were transfected with 0.5 µg of a Cas9-GFP construct (a gift from Kiran Musunuru, #44719, addgene, Cambridge, MA, USA), 0.5 µg gRNA expression vector, and 0.5 µg of dsRed expression vector using the Cell Line Nucleofector kit L (#VCA-1005, Lonza, Cologne, Germany). GFP/RFP double positive cells were single cell sorted 72 h post nucleofection, emerging cell clones were expanded, genotyped, and monitored for protein expression prior to phenotypic testing.
DNA binding in vitro
EMSAs were performed as described [9].
Formaldehyde-assisted isolation of regulatory elements (FAIRE)
FAIRE and calculation of the relative FAIRE enrichment was performed as described [8]. To purify DNA, the peqGOLD Cycle-Pure kit (011917, VWR, Darmstadt, Germany) was used according to the manufacturer’s protocol. For qPCR analyses, 40 ng of purified DNA served as template.
ChIP-qPCR, ChIP-seq, and data processing
ChIP-qPCR was performed as before [8]. For immunoprecipitation with histone antibodies, aliquots of 100 µg chromatin were used. Chromatin for ChIP-seq was prepared as described [8], except that cells were crosslinked for 5 min, and chromatin was sheared to 100–550 bp fragments by sonication for 12 min in a Covaris S220 device (Covaris, Woburn, MA, USA) with the following settings: peak incident power 150 W, duty factor 10%, and cycles/burst 200. After shearing, lysates were cleared by centrifugation (16 000 × g, 10 min, and 4 °C). The chromatin concentration in the supernatant was measured using a NanoDrop 2000 device (Thermo Fisher Scientific, Dreieich, Germany). Immunoprecipitations with 200 µg chromatin and 1 µg anti-HA antibody, and the subsequent washing steps were carried out as described [8]. To collect sufficient material for sequencing, 10–15 ChIPs were done in parallel. Samples were pooled before purifying immunoprecipitated DNA. DNA content was measured using the Qubit 2.0 system (Invitrogen, Paisley, UK). Two independent pools (=replicates) were prepared and submitted for library generation and subsequent 50 bp single-end sequencing on an Illumina HiSeq2000 (Illumina, San Diego, CA, USA). Bad quality reads were removed using trimmomatic [67]. Reads from ChIP and input samples were aligned to the human reference genome (hg19) with BWA aligner [68]. GATK was used for post processing analyses including local realignment and base quality score recalibration [69]. Peaks were called using the MACS2 software [70]. For each replicate, peaks were called on the aligned reads of ChIP samples using the corresponding input sample for background normalization. Peaks with an adjusted p-value below 0.05 were set as significant. The ChIPseeker R package was employed to assess the relation to closest gene and gene region annotations [71]. Bam files were converted to bigwig files using Galaxy and bamCoverage [72]. BigWig files were visualized with the UCSC genome browser [73]. ChIP-seq data files were deposited in GEO under the accession ID GSE127183.
Motif enrichment and GSEA
De novo motif analysis was performed using the rGADEM R packages [74]. The two ChIP-seq replicates were analyzed separately, an unseeded search was run with p-value and e-value parameters set to 0.0002 and zero, respectively. Motifs discovered were then mapped to known position weight matrices from the human transcription factor database [5] using MotIV R package [75]. The best match per motif was selected according to its e-value. GSEA of ChIP-seq peak-associated genes was examined separately for the two replicates using a hypergeometric test with the whole set of human protein-coding genes as background. GSEA was performed based on the Gene Ontology database “Biological Processes” and the ISC gene signature [21]. The Benjamini–Hochberg method was used for multiple-testing correction.
To compare different ChIP-seq data sets, Snail1-HA ChIP-seq replicates were merged using the “union” method from GenomicRanges R package [76]. For subsequent overlap analysis, Snail1-HA, ASCL2, and TCF7L2 ChIP-seq data sets were converted to hg38, using the UCSC liftover tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). For each gene region, we quantified the overlap between peak-associated genes in our data set and the published data sets [29, 77]. For Snail1 and Slug ChIP-seq data from murine cells, we filtered out peak-associated genes missing human homologs [10]. Significance of the overlap was assessed using a hypergeometric test with the sum of peak-associated genes in the tested sample as background.
Colony formation assays
For 2D colony formation assays, 1 × 103 cells/well were seeded in six-well plates in 2 ml DMEM with supplements. Medium, and if applicable Dox, was refreshed every 48 h. After 12 days of cultivation, medium was removed, cells were washed once with PBS and then stained with 1% crystal violet in 20% methanol for 10 min. Thereafter, the crystal violet solution was removed, cells were washed three times with H2O and then dried. Colonies were counted using the ImageJ software. For 3D colony formation assays, 2 × 103 cells/well were mixed with a top agar solution consisting of 0.7% sea plaque agarose (Lot 0000559478, Lonza, Basel, Switzerland) in DMEM with supplements. The cell suspension was seeded onto a base agar consisting of 1% sea plaque agarose in DMEM with supplements in 96-well plates. After solidification, the agar was overlaid with DMEM with supplements. Media and Dox were renewed every second day. Cells were cultured for 14 days, pictures were taken with a Nikon Eclipse TS100 microscope equipped with a Nikon DS-Qi1MC camera and colony numbers were determined. Colony diameter was measured using ImageJ software.
Limiting dilution assay
Cell numbers ranging from 1000 to 0.01 cells/well were seeded and cultivated in 96-well ultralow attachment plates. For each cell number, eight replicates were prepared. Media was refreshed every second day. Fourteen days after seeding the number of spheroid-containing wells per octaplicate were determined and subsequently processed using the ELDA tool (https//bioinf.wehi.edu.au/software/elda/) [40].
MTT assay
For MTT assays, 1 × 103 cells/well were seeded in 100 µl DMEM with supplements in 96-well plates. For each time point and condition, three wells were prepared as technical replicates. Where applicable, cells received 1 µg/ml Dox. Dox was refreshed after 48 h. 24 h (LS411 only) and 96 h after seeding, media were replaced by 20 µl of 5 mg/ml MTT in DMEM. Cells were incubated at 37 °C for 1 h, followed by removal of the supernatant. Cells were dissolved and the insoluble dye was extracted with 150 µl DMSO/well. The extinction of the solution was measured at 540 nm and 670 nm wavelength. For normalization, 670 nm values were subtracted from 540 nm values.
Flow cytometry
Cells were washed once with PBS and then trypsinized. Trypsinization was stopped by adding DMEM with supplements. Cells were centrifuged for 5 min at 150 × g, the supernatant was removed, cells were washed once with PBS, vortexed, and spun down again. The cells were then fixed in 70% ice-cold ethanol and stored at 4 °C. Prior to flow cytometry, the cell suspension was vortexed and then spun down at 150 × g. The supernatant was removed and the cell pellet was washed with 1× PBS. The washing step was repeated once before the cells were incubated with 20 µg/ml propidium iodide and 10 µg/ml RNAse in PBS for 30 min at 37 °C. Thereafter, the cell suspension was vortexed, spun down, washed with PBS, and resuspended in 200 µl to 400 µl PBS according to pellet size. Flow cytometric measurements were done with a CytoFlex S (Beckman Coulter, Indianapolis, USA).
Analysis of transcriptome data
Genes differentially expressed in the presence of Snail1-HA and bound by Snail1-HA, ASCL2, and TCF7L2, were identified upon processing the microarray data set GSE115716 as described [8]. Differentially expressed genes showing log2 fold changes of <−0.5 for downregulation, and >0.5 for upregulation (p-values < 0.05), respectively, were selected, and their association with ChIP-seq peaks for Snail1-HA, ASCL2, and TCF7L2 was analyzed at each time point post Snail1-HA induction. Note, that occupancy by Snail1-HA was determined only at t = 6 h. Kaplan–Meier plots were created using the XenaBrowser (http://xenabrowser.net). Studies selected for analyses were TCGA colon and rectal adenocarcinoma (COADREAD), BRCA, and pan-cancer (PANCAN). As gene-of-interest, MYB was picked. As assay type “IlluminaHiSeq gene expression RNAseq” was chosen. Samples were filtered for primary tumors. Pairwise-correlation analyses and calculation of relative gene expression levels based on CRC (GSE14333, GSE39582) and BRCA microarray data (TCGA BRCA) were performed as described [27, 28].
Statistics
Statistical analysis was performed using an unpaired, two-tailed Student’s t test. Similarity of variances was assessed by the F-test function implemented in GraphPad Prism v6.0. Unless otherwise stated, the comparison was between Dox-treated versus untreated cells and between parental versus genome-edited cells. Significant changes are shown by the respective p-values represented with *p < 0.05; **p < 0.01; ***p < 0.001. Non-significant changes: ns. Data are presented as mean + SEM. The sample size (=number of independent biological replicates) of each distinct experiment is indicated in the corresponding figure legend.
References
Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91.
Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28.
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018;172:650–65.
Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–9.
Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116(Pt 10):1959–67.
Jägle S, Busch H, Freihen V, Beyes S, Schrempp M, Boerries M, et al. SNAIL1-mediated downregulation of FOXA proteins facilitates the inactivation of transcriptional enhancer elements at key epithelial genes in colorectal cancer cells. PLoS Genet. 2017;13:e1007109.
Rönsch K, Jägle S, Rose K, Seidl M, Baumgartner F, Freihen V, et al. SNAIL1 combines competitive displacement of ASCL2 and epigenetic mechanisms to rapidly silence the EPHB3 tumor suppressor in colorectal cancer. Mol Oncol. 2015;9:335–54.
Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60.
Maturi V, Morén A, Enroth S, Heldin C-H, Moustakas A. Genomewide binding of transcription factor Snail1 in triple-negative breast cancer cells. Mol Oncol. 2018;12:1153–1174.
Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, Tajima K, et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 2013;5:1679–89.
Fabregat I, Malfettone A, Soukupova J. New insights into the crossroads between EMT and stemness in the context of cancer. J Clin Med. 2016;5:3.
Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15.
Morel A-P, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:e2888.
Preca B-T, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, et al. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer. 2015;137:2566–77.
Celià-Terrassa T, Meca-Cortés O, Mateo F, Martínez de Paz A, Rubio N, Arnal-Estapé A, et al. Epithelial–mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Investig. 2012;122:1849–68.
Caja L, Tzavlaki K, Dadras MS, Tan E-J, Hatem G, Maturi NP, et al. Snail regulates BMP and TGFβ pathways to control the differentiation status of glioma-initiating cells. Oncogene. 2018;37:2515–31.
Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11.
Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38.
Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo B-K, Itzkovitz S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–91.
van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–12.
Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–83.
Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7.
Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–9.
Batlle E, Henderson JT, Beghtel H, van den Born, Maaike MW, Sancho E, et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–63.
Schnappauf O, Beyes S, Dertmann A, Freihen V, Frey P, Jägle S, et al. Enhancer decommissioning by Snail1-induced competitive displacement of TCF7L2 and down-regulation of transcriptional activators results in EPHB2 silencing. Biochim Biophys Acta. 2016;1859:1353–67.
Jägle S, Rönsch K, Timme S, Andrlová H, Bertrand M, Jäger M, et al. Silencing of the EPHB3 tumor-suppressor gene in human colorectal cancer through decommissioning of a transcriptional enhancer. Proc Natl Acad Sci USA. 2014;111:4886–91.
Schuijers J, Junker JP, Mokry M, Hatzis P, Koo B-K, Sasselli V, et al. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–70.
van de Wetering M, Sancho E, Verweij C, Lau W, de, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50.
Haraguchi M, Okubo T, Miyashita Y, Miyamoto Y, Hayashi M, Crotti TN, et al. Snail regulates cell-matrix adhesion by regulation of the expression of integrins and basement membrane proteins. J Biol Chem. 2008;283:23514–23.
Tran DD, Corsa CAS, Biswas H, Aft RL, Longmore GD. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial–mesenchymal transition predicts for human breast cancer recurrence. Mol Cancer Res. 2011;9:1644–57.
Meyer-Schaller N, Cardner M, Diepenbruck M, Saxena M, Tiede S, Luond F, et al. A hierarchical regulatory landscape during the multiple stages of EMT. Dev Cell. 2019;48:539–553.e6.
Stanisavljevic J, Porta-de-la-Riva M, Batlle R, Herreros AG, Baulida J. The p65 subunit of NF-κB and PARP1 assist Snail1 in activating fibronectin transcription. J Cell Sci. 2011;124:4161–71. http://jcs.biologists.org/content/joces/124/24/4161.full.pdf.
Hsu DS-S, Wang H-J, Tai S-K, Chou C-H, Hsieh C-H, Chiu P-H, et al. Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell. 2014;26:534–48. http://www.sciencedirect.com/science/article/pii/S1535610814003651.
Rembold M, Ciglar L, Yáñez-Cuna JO, Zinzen RP, Girardot C, Jain A, et al. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014;28:167–81.
Giakountis A, Moulos P, Zarkou V, Oikonomou C, Harokopos V, Hatzigeorgiou AG, et al. A positive regulatory loop between a Wnt-regulated non-coding RNA and ASCL2 controls intestinal stem cell fate. Cell Rep. 2016;15:2588–96.
Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–42.
Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85.
Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8.
Stemmer V, de Craene B, Berx G, Behrens J. Snail promotes Wnt target gene expression and interacts with beta-catenin. Oncogene. 2008;27:5075–80.
Rosmaninho P, Mukusch S, Piscopo V, Teixeira V, Raposo AA, Warta R. et al. Zeb1 potentiates genome-wide gene transcription with Lef1 to promote glioblastoma cell invasion. EMBO J. 2018;37:e97115.
Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CEM, van Ijcken, et al. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell. 2012;47:457–68.
Millanes-Romero A, Herranz N, Perrera V, Iturbide A, Loubat-Casanovas J, Gil J, et al. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol Cell. 2013;52:746–57.
Lin Y, Wu Y, Li J, Dong C, Ye X, Chi Y-I, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–16.
Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–19.
Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–34.
Malaterre J, Pereira L, Putoczki T, Millen R, Paquet-Fifield S, Germann M, et al. Intestinal-specific activatable Myb initiates colon tumorigenesis in mice. Oncogene. 2016;35:2475–84.
Malaterre J, Carpinelli M, Ernst M, Alexander W, Cooke M, Sutton S, et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc Natl Acad Sci USA. 2007;104:3829–34.
Srivastava SK, Bhardwaj A, Arora S, Singh S, Azim S, Tyagi N, et al. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br J Cancer. 2015;113:1694–703.
Miao RY, Drabsch Y, Cross RS, Cheasley D, Carpinteri S, Pereira L, et al. MYB is essential for mammary tumorigenesis. Cancer Res. 2011;71:7029–37.
Ciznadija D, Tothill R, Waterman ML, Zhao L, Huynh D, Yu RM, et al. Intestinal adenoma formation and MYC activation are regulated by cooperation between MYB and Wnt signaling. Cell Death Differ. 2009;16:1530–8.
Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43.
Hugo HJ, Pereira L, Suryadinata R, Drabsch Y, Gonda TJ, Gunasinghe NPA Devika. et al. Direct repression of MYB by ZEB1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res. 2013;15:R113
Guinney J, Dienstmann R, Wang X, Reyniès A, de, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6.
Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;1134. https://www.nature.com/articles/s41556-018-0194-0.pdf.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5.
Cheasley D, Pereira L, Lightowler S, Vincan E, Malaterre J, Ramsay RG. Myb controls intestinal stem cell genes and self-renewal. Stem Cells. 2011;29:2042–50.
Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79.
Hwang W-L, Jiang J-K, Yang S-H, Huang T-S, Lan H-Y, Teng H-W, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–80.
Hwang W-L, Yang M-H, Tsai M-L, Lan H-Y, Su S-H, Chang S-C, et al. SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology. 2011;141:279–91.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56.
Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wöhrle S, et al. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964–81.
Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–95.
Van der Auwera, Geraldine A, Carneiro MO, Hartl C, Poplin R, Del Angel G, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–33.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
rGADEM: Bioconductor. 2017. https://doi.org/10.18129/B9.BIOC.RGADEM.
MotIV: Bioconductor. 2017. https://doi.org/10.18129/B9.BIOC.MOTIV.
Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118.
Mokry M, Hatzis P, de Bruijn E, Koster J, Versteeg R, Schuijers J, et al. Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PLoS ONE. 2010;5:e15092.
Acknowledgements
The authors are grateful to K. Geiger and D. Herchenbach for excellent technical assistance, and to the team of the Genomics and Proteomics Core Facility, German Cancer Research Center/DKFZ, Heidelberg, Germany, for their sequencing service. This work was financially supported by the Deutsche Forschungsgemeinschaft (DFG HE2004/11-1 to AH; CRC-850 subprojects B5 to AH and Z1 to MB; GRK2344 subprojects B02 to MB and B03 to AH), and by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept CoNfirm (FKZ 01ZX1708F to MB). JW and PA-G were funded by the Deutsche Forschungsgemeinschaft Research Training Group GRK2344 “MeInBio—BioInMe”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Beyes, S., Andrieux, G., Schrempp, M. et al. Genome-wide mapping of DNA-binding sites identifies stemness-related genes as directly repressed targets of SNAIL1 in colorectal cancer cells. Oncogene 38, 6647–6661 (2019). https://doi.org/10.1038/s41388-019-0905-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0905-4
This article is cited by
-
SMAD4 mutations do not preclude epithelial–mesenchymal transition in colorectal cancer
Oncogene (2022)
-
Loss of SNAI1 induces cellular plasticity in invasive triple-negative breast cancer cells
Cell Death & Disease (2022)
-
Canonical TGFβ signaling induces collective invasion in colorectal carcinogenesis through a Snail1- and Zeb1-independent partial EMT
Oncogene (2022)
-
Transcription factor c-Myb: novel prognostic factor in osteosarcoma
Clinical & Experimental Metastasis (2022)
-
Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer
Journal of Translational Medicine (2021)