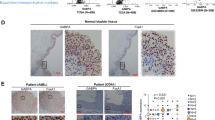
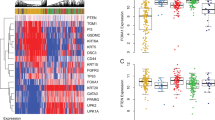
Abstract
Basal subtype cancers are deadly malignancies but the molecular events driving tumor lethality are not completely understood. Ataxia-telangiectasia group D complementing gene (ATDC, also known as TRIM29), is highly expressed and drives tumor formation and invasion in human bladder cancers but the factor(s) regulating its expression in bladder cancer are unknown. Molecular subtyping of bladder cancer has identified an aggressive basal subtype, which shares molecular features of basal/squamous tumors arising in other organs and is defined by activation of a TP63-driven gene program. Here, we demonstrate that ATDC is linked with expression of TP63 and highly expressed in basal bladder cancers. We find that TP63 binds to transcriptional regulatory regions of ATDC and KRT14 directly, increasing their expression, and that ATDC and KRT14 execute a TP63-driven invasive program. In vivo, ATDC is required for TP63-induced bladder tumor invasion and metastasis. These results link TP63 and the basal gene expression program to ATDC and to aggressive tumor behavior. Defining ATDC as a molecular determinant of aggressive, basal cancers may lead to improved biomarkers and therapeutic approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. https://doi.org/10.3322/caac.21332.
Network, T. C. G. A. R. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature https://doi.org/10.1038/nature12965 (2014).
Damrauer JS, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci USA. 2014;111:3110–5. https://doi.org/10.1073/pnas.1318376111.
Choi W, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65. https://doi.org/10.1016/j.ccr.2014.01.009.
Hedegaard J, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30:27–42. https://doi.org/10.1016/j.ccell.2016.05.004.
Sjödahl G, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86. https://doi.org/10.1158/1078-0432.CCR-12-0077-T.
Lerner SP, et al. Bladder cancer molecular taxonomy: summary from a consensus meeting. Bladder Cancer. 2016;2:37–47. https://doi.org/10.3233/BLC-150037.
Choi W, et al. Genetic alterations in the molecular subtypes of bladder cancer: illustration in the cancer genome atlas dataset. Eur Urol. 2017;72:354–65. https://doi.org/10.1016/j.eururo.2017.03.010.
Robertson AG, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540 https://doi.org/10.1016/j.cell.2017.09.007. e525.
Sjodahl G, Eriksson P, Liedberg F, Hoglund M. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J Pathol. 2017;242:113–25. https://doi.org/10.1002/path.4886.
Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. https://doi.org/10.1016/j.cell.2014.06.049.
Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–72. https://doi.org/10.1038/sj.cdd.4401914.
Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer. 2013;13:136–43. https://doi.org/10.1038/nrc3446.
Karni-Schmidt O, et al. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am J Pathol. 2011;178:1350–60. https://doi.org/10.1016/j.ajpath.2010.11.061.
Lai W, et al. Upregulated ataxia-telangiectasia group D complementing gene correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Dis Esophagus. 2013;26:817–22. https://doi.org/10.1111/j.1442-2050.2012.01400.x.
Palmbos PL, et al. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Cancer Res. 2015;75:5155–66. https://doi.org/10.1158/0008-5472.CAN-15-0603.
Tang ZP, et al. Ataxia-telangiectasia Group D complementing gene (ATDC) promotes lung cancer cell proliferation by activating NF-κB pathway. PLoS ONE. 2013;8:e63676. https://doi.org/10.1371/journal.pone.0063676.
Wang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15:207–19. S1535-6108(09)00027-0 [pii] 10.1016/j.ccr.2009.01.018.
Wang L, et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. 2015;29:171–83. https://doi.org/10.1101/gad.253591.114.
Qiu F, Xiong JP, Deng J, Xiang XJ. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. 2015;8:5053–61.
Dükel M, et al. The breast cancer tumor suppressor TRIM29 is expressed via ATM-dependent signaling in response to hypoxia. J Biol Chem. 2016;291:21541–52. https://doi.org/10.1074/jbc.M116.730960.
Guo GC, Wang JX, Han ML, Zhang LP, Li L. microRNA-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression. Cell Oncol. 2017;40:157–66. https://doi.org/10.1007/s13402-016-0312-6.
Guo, G et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. https://doi.org/10.1038/ng.2798 (2013).
Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. https://doi.org/10.1158/2159-8290.CD-12-0095.
Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1 https://doi.org/10.1126/scisignal.2004088.
Tamura S. et al. Molecular correlates of in vitro responses to dacomitinib and afatinib in bladder cancer. Bladder Cancer. 2018;4:77–90.
Hovelson, DH et al. Targeted DNA and RNA sequencing of paired urothelial and squamous bladder cancers reveals discordant genomic and transcriptomic events and unique therapeutic implications. Eur Urol. https://doi.org/10.1016/j.eururo.2018.06.047 (2018).
Choi W, et al. p63 expression defines a lethal subset of muscle-invasive bladder cancers. PLoS ONE. 2012;7:e30206 https://doi.org/10.1371/journal.pone.0030206.
McDade SS, et al. Genome-wide analysis of p63 binding sites identifies AP-2 factors as co-regulators of epidermal differentiation. Nucleic Acids Res. 2012;40:7190–206. https://doi.org/10.1093/nar/gks389.
Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. https://doi.org/10.1101/gr.229102.. Article published online before print in May 2002.
Candi E, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–47. https://doi.org/10.1038/sj.cdd.4401926.
Masuda Y, Takahashi H, Hatakeyama S. TRIM29 regulates the p63-mediated pathway in cervical cancer cells. Biochim Biophys Acta. 2015;1853:2296–305. https://doi.org/10.1016/j.bbamcr.2015.05.035.
Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155:1639–51. https://doi.org/10.1016/j.cell.2013.11.029.
Wang, F, Chen, S, Liu, HB, Parent, CA, Coulombe. PA Keratin 6 regulates collective keratinocyte migration by altering cell–cell and cell–matrix adhesion. J Cell Biol. https://doi.org/10.1083/jcb.201712130 (2018).
Velez-delValle C, Marsch-Moreno M, Castro-Munozledo F, Galvan-Mendoza IJ, Kuri-Harcuch W. Epithelial cell migration requires the interaction between the vimentin and keratin intermediate filaments. Sci Rep. 2016;6:24389 https://doi.org/10.1038/srep24389.
Xu W, et al. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol Rep. 2016;36:1411–8. https://doi.org/10.3892/or.2016.4941.
Wang, Y, Day, ML, Simeone, DM, Palmbos, PL. 3-D cell culture system for studying invasion and evaluating therapeutics in bladder cancer. J Vis Exp. https://doi.org/10.3791/58345 (2018).
Seiler R, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017. https://doi.org/10.1016/j.eururo.2017.03.030.
Lee SH, et al. The Dishevelled-binding protein CXXC5 negatively regulates cutaneous wound healing. J Exp Med. 2015;212:1061–80. https://doi.org/10.1084/jem.20141601.
Yuan Z, et al. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. 2010;30:3004–15. MCB.01023-09 [pii] 10.1128/MCB.01023-09.
Cai BH, et al. p53 acts as a co-repressor to regulate keratin 14 expression during epidermal cell differentiation. PLoS ONE. 2012;7:e41742. https://doi.org/10.1371/journal.pone.0041742.
Leivo MZ, Elson PJ, Tacha DE, Delahunt B, Hansel DE. A combination of p40, GATA-3 and uroplakin II shows utility in the diagnosis and prognosis of muscle-invasive urothelial carcinoma. Pathology. 2016;48:543–9. https://doi.org/10.1016/j.pathol.2016.05.008.
Gaya JM, et al. ΔNp63 expression is a protective factor of progression in clinical high grade T1 bladder cancer. J Urol. 2015;193:1144–50. https://doi.org/10.1016/j.juro.2014.10.098.
Han AL, et al. Fibulin-3 promotes muscle-invasive bladder cancer. Oncogene. 2017. https://doi.org/10.1038/onc.2017.149.
Acknowledgments
The authors would like to thank Andrew Ewald for assistance with the 3D invasion and imaging systems used in this study.
Funding
This work was funded by grants from the University of Michigan Cancer Center Core Grant CA046592-26S3, NIH K08 CA201335 (PLP), BCAN YIA (PLP), ASCO YIA (PLP), and NIH R01 CA17483601 (DMS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Palmbos, P.L., Wang, Y., Bankhead III, A. et al. ATDC mediates a TP63-regulated basal cancer invasive program. Oncogene 38, 3340–3354 (2019). https://doi.org/10.1038/s41388-018-0646-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0646-9
This article is cited by
-
TP63–TRIM29 axis regulates enhancer methylation and chromosomal instability in prostate cancer
Epigenetics & Chromatin (2024)
-
Subtype-associated epigenomic landscape and 3D genome structure in bladder cancer
Genome Biology (2021)
-
Repression of transcription factor AP-2 alpha by PPARγ reveals a novel transcriptional circuit in basal-squamous bladder cancer
Oncogenesis (2019)