Abstract
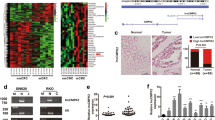
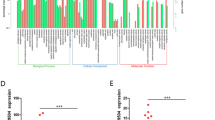
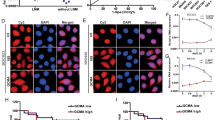
Long noncoding RNAs (lncRNAs) are pervasive transcripts that play pivotal roles in regulating chromatin dynamics, gene and protein expression. Aberrant expression and mutations of lncRNAs represent a driving force behind tumor invasion and metastasis, making them attractive cancer targets. However, most of the lncRNAs are still being discovered and conclusive experimental evidence for their functional relevance is still lacking for most malignancies. In this study, a differentially expressed lncRNA, designated as lnc-CRCMSL, is identified by microarray-based screenings on non-metastatic and metastatic CRC specimens. Lnc-CRCMSL is verified as an anti-metastatic gene and negatively correlated with the poor prognosis of CRC patients. Lnc-CRCMSL overexpression restricts tumor growth and metastasis in vivo and in vitro. Instead, lnc-CRCMSL silencing accelerates CRC cell proliferation and migration. RNA-pulldown assay identifies high mobility group box 2 (HMGB2) as a downstream protein of lnc-CRCMSL. Mechanically, lnc-CRCMSL physically binds to HMGB2 and stabilizes the localization of HMGB2 in the cytoplasm. Notably, lnc-CRCMSL knockdown lead to the shift of HMGB2 into nuclear, in which it triggers epithelial to mesenchymal transition (EMT) programming. Importantly, lnc-CRCMSL controls the cytoplasmic retention of HMGB2 and attenuates the interaction between HMGB2 and OCT4 to suppress EMT. Treatment of leptomycin B (LMB), a potent and specific nuclear export inhibitor, counteracts lnc-CRCMSL-mediated suppression of aggressive phenotypes and EMT process by accumulating the nuclear HMGB2.
Conclusion
Our data highlight the anti-metastatic role of lnc-CRCMSL in stabilizing HMGB2 through lncRNA-protein interactions in the cytoplasm, and suggest that targeting lnc-CRCMSL may represent a therapeutic opportunity for managing metastatic CRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Perez-Lluch S, Abad A, Davis C, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–40.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.
Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long Non-coding RNAs and their role in metastasis. Cancer Genom Proteom. 2017;14:143–60.
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–26.
Huang Q, Yan J, Agami R. Long non-coding RNAs in metastasis. Cancer Metastas Rev. 2018;37:75–81.
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61.
Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–7.
Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–516.
Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang F, et al. LIM and SH3 protein 1 induces TGFbeta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res. 2014;20:5835–47.
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603.
Zhang Y, Wagner EK, Guo X, May I, Cai Q, Zheng W, et al. Long intergenic non-coding RNA expression signature in human breast cancer. Sci Rep. 2016;6:37821.
Wang P, Cui J, Wen J, Guo Y, Zhang L, Chen X. Cisplatin induces HepG2 cell cycle arrest through targeting specific long noncoding RNAs and the p53 signaling pathway. Oncol Lett. 2016;12:4605–12.
Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang S, et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 2010;59:1226–35.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21.
Pennisi E. Long noncoding RNAs may alter chromosome’s 3D structure. Science. 2013;340:910.
Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair. 2015;36:122–36.
Takeuchi T, Sakazume K, Tonooka A, Zaitsu M, Takeshima Y, Mikami K, et al. Cytosolic HMGB1 expression in human renal clear cell cancer indicates higher pathological T classifications and tumor grades. Urol J. 2013;10:960–5.
Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochim Biophys Acta. 2010;1799:114–8.
Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5.
Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16:5511–21.
Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal. 2018;16:8.
Castillo-Quan JI, Blackwell TK. Metformin: restraining nucleocytoplasmic shuttling to fight cancer and aging. Cell. 2016;167:1670–1.
Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, et al. MicroRNA-187, a downstream effector of TGFbeta pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–13.
Islam SS, Mokhtari RB, Noman AS, Uddin M, Rahman MZ, Azadi MA, et al. Sonic hedgehog (Shh) signaling promotes tumorigenicity and stemness via activation of epithelial-to-mesenchymal transition (EMT) in bladder cancer. Mol Carcinog. 2016;55:537–51.
Aird KM, Iwasaki O, Kossenkov AV, Tanizawa H, Fatkhutdinov N, Bitler BG, et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J Cell Biol. 2016;215:325–34.
Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–7.
Campbell PA, Rudnicki MA. Oct4 interaction with Hmgb2 regulates Akt signaling and pluripotency. Stem Cells. 2013;31:1107–20.
Mandal G, Biswas S, Chowdhury SR, Chatterjee A, Purohit S, Khamaru P, et al. Heterodimer formation by Oct4 and Smad3 differentially regulates epithelial-to-mesenchymal transition-associated factors in breast cancer progression. Biochim Biophys Acta. 2018;1864:2053–66.
Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, et al. betaII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology. 2015;61:598–612.
Zhao Y, Yang Z, Wu J, Wu R, Keshipeddy SK, Wright D, et al. High-mobility-group protein 2 regulated by microRNA-127 and small heterodimer partner modulates pluripotency of mouse embryonic stem cells and liver tumor initiating cells. Hepatol Commun. 2017;1:816–30.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–14.
Zhang YF, Liu L, Ding YQ. [Isolation and characterization of human colorectal cancer cell subline with unique metastatic potential in the liver]. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:126–30.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81572813, 81773082, 81573848, 81774172), Guangdong Natural Science Foundation (2015A030313274), Science and Technology Program of Guangzhou (1563000235) and Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (pdjhb0105).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Precis: Findings identify and report a CRC metastasis suppressor lnc-CRCMSL with potential therapeutic applications.
Supplementary information
Rights and permissions
About this article
Cite this article
Han, Q., Xu, L., Lin, W. et al. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene 38, 3019–3032 (2019). https://doi.org/10.1038/s41388-018-0614-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0614-4
This article is cited by
-
Sirt3 restricts tumor initiation via promoting LONP1 deacetylation and K63 ubiquitination
Journal of Translational Medicine (2023)
-
Long non-coding RNAs as the critical regulators of epithelial mesenchymal transition in colorectal tumor cells: an overview
Cancer Cell International (2022)
-
The LncRNA RP11-301G19.1/miR-582-5p/HMGB2 axis modulates the proliferation and apoptosis of multiple myeloma cancer cells via the PI3K/AKT signalling pathway
Cancer Gene Therapy (2022)
-
Long Non-coding RNAs (lncRNAs), A New Target in Stroke
Cellular and Molecular Neurobiology (2022)
-
NF-κB-activated SPRY4-IT1 promotes cancer cell metastasis by downregulating TCEB1 mRNA via Staufen1-mediated mRNA decay
Oncogene (2021)