Abstract
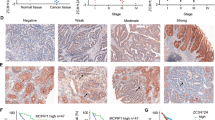
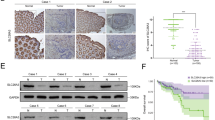
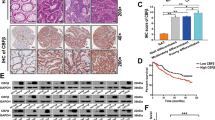
Multiple studies have shown that chronic inflammation is closely related to the occurrence and development of colorectal cancer (CRC). Classical NF-κB signaling, the key factor in controlling inflammation, has been found to be of great importance to CRC development. However, the role of alternative NF-κB signaling in CRC is still elusive. Here, we found aberrant constitutive activation of alternative NF-κB signaling both in CRC tissue and CRC cells. Knockdown of RelB downregulates c-Myc and upregulates p27Kip1 protein level, which inhibits CRC cell proliferation and retards CRC xenograft growth. Conversely, overexpression of RelB increases proliferation of CRC cells. In addition, we revealed a significant correlation between Bcl-3 and RelB in CRC tissues. The expression of RelB was consistent with the expression of Bcl-3 and the phosphorylation of Bcl-3 downstream proteins p-Akt (S473) and p-GSK3β (S9). Bcl-3 overexpression can restore the phenotype changes caused by RelB knockdown. Importantly, we demonstrated that alternative NF-κB transcriptional factor (p52:RelB) can directly bind to the promoter region of Bcl-3 gene and upregulate its transcription. Moreover, the expression of RelB, NF-κB2 p52, and Bcl-3 was associated with poor survival of CRC patients. Taken together, these results represent that alternative NF-κB signaling may function as an oncogenic driver in CRC, and also provide new ideas and research directions for the pathogenesis, prevention, and treatment of other inflammatory-related diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30.
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–93.
Karin M, NF-kB and cancer: mechanisms and targets. Mol Carcinog. 2006;45:355–61.
Ghosh MSHS. Shared principles in NF-κB signaling. Cell. 2008;132:344–62.
Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–14 e5.
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H, Hayakawa Y, Shibata W, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15:2248–58.
Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–54.
Dejardin E, Deregowski V, Chapelier M, Jacobs N, Gielen J, Merville MP, et al. Regulation of NF-kappaB activity by I kappaB-related proteins in adenocarcinoma cells. Oncogene. 1999;18:2567–77.
Lessard L, Begin LR, Gleave ME, Mes-Masson AM, Saad F. Nuclear localisation of nuclear factor-kappaB transcription factors in prostate cancer: an immunohistochemical study. Br J Cancer. 2005;93:1019–23.
Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30.
Wharry CE, Haines KM, Carroll RG, May MJ. Constitutive non-canonical NFkappaB signaling in pancreatic cancer cells. Cancer Biol Ther. 2009;8:1567–76.
Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci USA. 2011;108:14596–601.
Jacque E, Billot K, Authier H, Bordereaux D, Baud V. RelB inhibits cell proliferation and tumor growth through p53 transcriptional activation. Oncogene. 2013;32:2661–9.
Xu Y, Josson S, Fang F, Oberley TD, St Clair DK, Wan XS, et al. RelB enhances prostate cancer growth: implications for the role of the nuclear factor-kappaB alternative pathway in tumorigenicity. Cancer Res. 2009;69:3267–71.
Nottingham LK, Yan CH, Yang X, Si H, Coupar J, Bian Y, et al. Aberrant IKKalpha and IKKbeta cooperatively activate NF-kappaB and induce EGFR/AP1 signaling to promote survival and migration of head and neck cancer. Oncogene. 2014;33:1135–47.
Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, et al. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat Cell Biol. 2007;9:470–8.
Ge QL, Liu SH, Ai ZH, Tao MF, Ma L, Wen SY, et al. RelB/NF-kappaB links cell cycle transition and apoptosis to endometrioid adenocarcinoma tumorigenesis. Cell Death Dis. 2016;7:e2402.
Vallabhapurapu SD, Noothi SK, Pullum DA, Lawrie CH, Pallapati R, Potluri V, et al. Transcriptional repression by the HDAC4-RelB-p52 complex regulates multiple myeloma survival and growth. Nat Commun. 2015;6:8428.
Ohno H, Takimoto G, McKeithan TW. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60:991–7.
Bours V, Villalobos J, Burd PR, Kelly K, Siebenlist U. Cloning of a mitogen-inducible gene encoding a kappa B DNA-binding protein with homology to the rel oncogene and to cell-cycle motifs. Nature. 1990;348:76–80.
Tanaka S, Nishigaki H, Nakagawa H, Okuda T, Nishida K, Tsuda S, et al. Reciprocal t(14;19)(q32.3; q13.1) in a patient with B-cell lymphoma. Cancer Genet Cytogenet. 1990;49:219–24.
Naumann M, Wulczyn FG, Scheidereit C. The NF-kappa B precursor p105 and the proto-oncogene product Bcl-3 are I kappa B molecules and control nuclear translocation of NF-kappa B. EMBO J. 1993;12:213–22.
Nolan GP, Fujita T, Bhatia K, Huppi C, Liou HC, Scott ML, et al. The bcl-3 proto-oncogene encodes a nuclear I kappa B-like molecule that preferentially interacts with NF-kappa B p50 and p52 in a phosphorylation-dependent manner. Mol Cell Biol. 1993;13:3557–66.
Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, et al. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–39.
Maldonado V, Melendez-Zajgla J. Role of Bcl-3 in solid tumors. Mol Cancer. 2011;10:152.
Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y, et al. The IkappaB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 2013;5:280–2.
Urban BC, Collard TJ, Eagle CJ, Southern SL, Greenhough A, Hamdollah-Zadeh M, et al. BCL-3 expression promotes colorectal tumorigenesis through activation of AKT signalling. Gut. 2016;65:1151–64.
Brasier AR, Lu M, Hai T, Lu Y, Boldogh I. NF-kappa B-inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-kappa B1 residence. J Biol Chem. 2001;276:32080–93.
Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–59 e11.
Wang Y, Xu J, Gao G, Li J, Huang H, Jin H, et al. Tumor-suppressor NFkappaB2p100 interacts with ERK2 and stabilizes PTEN mRNA via inhibition of miR-494. Oncogene. 2016;35:4080–90.
Muller JR, Siebenlist U. Lymphotoxin beta receptor induces sequential activation of distinct NF-kappa B factors via separate signaling pathways. J Biol Chem. 2003;278:12006–12.
Wang S, Liu Z, Wang L, Zhang X. NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6:327–34.
Wen D, Nong Y, Morgan JG, Gangurde P, Bielecki A, Dasilva J, et al. A selective small molecule IkappaB Kinase beta inhibitor blocks nuclear factor kappaB-mediated inflammatory responses in human fibroblast-like synoviocytes, chondrocytes, and mast cells. J Pharmacol Exp Ther. 2006;317:989–1001.
Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Schumm K, Rocha S, Caamano J, Perkins ND. Regulation of p53 tumour suppressor target gene expression by the p52 NF-kappaB subunit. EMBO J. 2006;25:4820–32.
Zhang J, Warren MA, Shoemaker SF, Ip MM. NFkappaB1/p50 is not required for tumor necrosis factor-stimulated growth of primary mammary epithelial cells: implications for NFkappaB2/p52 and RelB. Endocrinology. 2007;148:268–78.
Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90.
Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:d250–68.
Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, et al. c-Myc represses FOXO3a-mediated transcription of the gene encoding thep27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem. 2008;104:2091–106.
Perez-Roger I, Solomon DL, Sewing A, Land H. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition ofp27(Kip1) binding to newly formed complexes. Oncogene. 1997;14:2373–81.
Bretones G, Acosta JC, Caraballo JM, Ferrandiz N, Gomez-Casares MT, Albajar M, et al. SKP2 oncogene is a direct MYC target gene and MYC down-regulatesp27(KIP1) through SKP2 in human leukemia cells. J Biol Chem. 2011;286:9815–25.
Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT, Mei Q, et al. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2alpha and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene. 2015;34:1393–406.
Chen C, Cai S, Wang G, Cao X, Yang X, Luo X, et al. c-Myc enhances colon cancer cell-mediated angiogenesis through the regulation of HIF-1alpha. Biochem Biophys Res Commun. 2013;430:505–11.
Saamarthy K, Bjorner S, Johansson M, Landberg G, Massoumi R, Jirstrom K, et al. Early diagnostic value of Bcl-3 localization in colorectal cancer. BMC Cancer. 2015;15:341.
Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J Exp Med. 1998;187:147–59.
Weih F, Caamano J. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol Rev. 2003;195:91–105.
Poljak L, Carlson L, Cunningham K, Kosco-Vilbois MH, Siebenlist U. Distinct activities of p52/NF-kappa B required for proper secondary lymphoid organ microarchitecture: functions enhanced by Bcl-3. J Immunol. 1999;163:6581–8.
Zhang X, Wang H, Claudio E, Brown K, Siebenlist U. A role for the IkappaB family member Bcl-3 in the control of central immunologic tolerance. Immunity. 2007;27:438–52.
Franzoso G, Carlson L, Scharton-Kersten T, Shores EW, Epstein S, Grinberg A, et al. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture, and germinal center reactions. Immunity. 1997;6:479–90.
Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–35.
Acknowledgements
This work was supported by the National Program on Key Research (2016YFC1302400), the National Basic Research Program (2014CB541904, 2014CB943600), National Natural Science Foundation of China (91742113, 31570902, 31370881), and Natural Science Foundation of Shanghai (14ZR1426300, 18ZR1424400, 18ZR1446400).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Yu Tao, Zhanjie Liu, Yingyong Hou.
Rights and permissions
About this article
Cite this article
Tao, Y., Liu, Z., Hou, Y. et al. Alternative NF-κB signaling promotes colorectal tumorigenesis through transcriptionally upregulating Bcl-3. Oncogene 37, 5887–5900 (2018). https://doi.org/10.1038/s41388-018-0363-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0363-4
This article is cited by
-
Multifaceted roles for BCL3 in cancer: a proto-oncogene comes of age
Molecular Cancer (2024)
-
Aberrant non-canonical NF-κB signalling reprograms the epigenome landscape to drive oncogenic transcriptomes in multiple myeloma
Nature Communications (2024)
-
Multiple roles for Bcl-3 in mammary gland branching, stromal collagen invasion, involution and tumor pathology
Breast Cancer Research (2022)
-
OLFM4 deficiency delays the progression of colitis to colorectal cancer by abrogating PMN-MDSCs recruitment
Oncogene (2022)
-
The hepatic senescence-associated secretory phenotype promotes hepatocarcinogenesis through Bcl3-dependent activation of macrophages
Cell & Bioscience (2021)