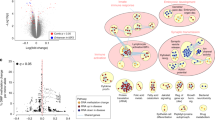
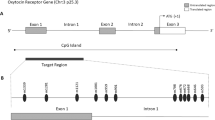
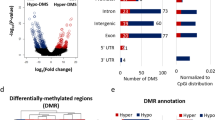
Abstract
Introduction
A microdeletion including the SNORD116 gene (SNORD116 MD) has been shown to drive the Prader-Willi syndrome (PWS) features. PWS is a neurodevelopmental disorder clinically characterized by endocrine impairment, intellectual disability and psychiatric symptoms such as a lack of emotional regulation, impulsivity, and intense temper tantrums with outbursts. In addition, this syndrome is associated with a nutritional trajectory characterized by addiction-like behavior around food in adulthood. PWS is related to the genetic loss of expression of a minimal region that plays a potential role in epigenetic regulation. Nevertheless, the role of the SNORD116 MD in DNA methylation, as well as the impact of the oxytocin (OXT) on it, have never been investigated in human neurons.
Methods
We studied the methylation marks in induced pluripotent stem-derived dopaminergic neurons carrying a SNORD116 MD in comparison with those from an age-matched adult healthy control. We also performed identical neuron differentiation in the presence of OXT. We performed a genome-wide DNA methylation analysis from the iPSC-derived dopaminergic neurons by reduced-representation bisulfite sequencing. In addition, we performed RNA sequencing analysis in these iPSC-derived dopaminergic neurons differentiated with or without OXT.
Results
The analysis revealed that 153,826 cytosines were differentially methylated between SNORD116 MD neurons and control neurons. Among the differentially methylated genes, we determined a list of genes also differentially expressed. Enrichment analysis of this list encompassed the dopaminergic system with COMT and SLC6A3. COMT displayed hypermethylation and under-expression in SNORD116 MD, and SLC6A3 displayed hypomethylation and over-expression in SNORD116 MD. RT-qPCR confirmed significant over-expression of SLC6A3 in SNORD116 MD neurons. Moreover, the expression of this gene was significantly decreased in the case of OXT adjunction during the differentiation.
Conclusion
SNORD116 MD dopaminergic neurons displayed differential methylation and expression in the COMT and SLC6A3 genes, which are related to dopaminergic clearance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA sequencing data reported in this article were deposited in the NCBI’s Gene Expression Omnibus (GEO) database: GSE249891.
References
Tauber M, Diene G, Mimoun E, Çabal-Berthoumieu S, Mantoulan C, Molinas C, et al. Prader-Willi syndrome as a model of human hyperphagia. Front Horm Res. 2014;42:93–106.
McCandless SE, The Committee on Genetics. Health supervision for children with Prader-Willi syndrome. Pediatrics. 2011;127:195–204.
Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29:299–308.
Edwards AC, Maes HH, Pedersen NL, Kendler KS. A population-based twin study of the genetic and environmental relationship of major depression, regular tobacco use and nicotine dependence. Psychol Med. 2011;41:395–405.
Edwards AC, Kendler KS. A twin study of depression and nicotine dependence: shared liability or causal relationship? J Affect Disord. 2012;142:90–7.
Bierut LJ. Nicotine dependence and genetic variation in the nicotinic receptors. Drug Alcohol Depend. 2009;104:S64–9.
Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. JAMA. 1992;268:1877–82.
Heath AC, Whitfield JB, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, et al. Towards a molecular epidemiology of alcohol dependence: analysing the interplay of genetic and environmental risk factors. Br J Psychiatry Suppl. 2001;40:s33–40.
Heath AC, Martin NG. Genetic influences on alcohol consumption patterns and problem drinking: results from the Australian NH&MRC twin panel follow-up survey. Ann N Y Acad Sci. 1994;708:72–85.
Slutske WS, Zhu G, Meier MH, Martin NG. Genetic and environmental influences on disordered gambling in men and women. Arch Gen Psychiatry. 2010;67:624–30.
Blum K, Braverman ER, Holder JM, Lubar JF, Monastra VJ, Miller D, et al. Reward deficiency syndrome: a biogenetic model for the diagnosis and treatment of impulsive, addictive, and compulsive behaviors. J Psychoact Drugs. 2000;32:1–112.
Lindgren E, Gray K, Miller G, Tyler R, Wiers CE, Volkow ND, et al. Food addiction: a common neurobiological mechanism with drug abuse. Front Biosci Landmark Ed. 2018;23:811–36.
Wang SC, Chen YC, Lee CH, Cheng CM. Opioid addiction, genetic susceptibility, and medical treatments: a review. Int J Mol Sci. 2019;20:4294.
Volkow ND, Koob GF, McLellan AT. Neurobiologic advances from the brain disease model of addiction. N Engl J Med. 2016;374:363–71.
Ross S, Peselow E. Co-occurring psychotic and addictive disorders: neurobiology and diagnosis. Clin Neuropharmacol. 2012;35:235–43.
Maldonado R, Calvé P, García-Blanco A, Domingo-Rodriguez L, Senabre E, Martín-García E. Genomics and epigenomics of addiction. Am J Med Genet B Neuropsychiatr Genet. 2021;186:128–39.
Mancino S, Burokas A, Gutiérrez-Cuesta J, Gutiérrez-Martos M, Martín-García E, Pucci M, et al. Epigenetic and proteomic expression changes promoted by eating addictive-like behavior. Neuropsychopharmacology. 2015;40:2788–800.
Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, et al. DNA methylation regulates associative reward learning. Nat Neurosci. 2013;16:1445–52.
Strathearn L. Maternal neglect: oxytocin, dopamine and the neurobiology of attachment. J Neuroendocrinol. 2011;23:1054–65.
Insel TR, Young LJ. The neurobiology of attachment. Nat Rev Neurosci. 2001;2:129–36.
Carter CS. The role of oxytocin and vasopressin in attachment. Psychodyn Psychiatry. 2017;45:499–517.
Pedersen CA. Oxytocin, tolerance, and the dark side of addiction. Int Rev Neurobiol. 2017;136:239–74.
Whittington J, Holland A. Cognition in people with Prader-Willi syndrome: insights into genetic influences on cognitive and social development. Neurosci Biobehav Rev. 2017;72:153–67.
Viaux-Savelon S, Rosenblum O, Guedeney A, Diene G, Çabal-Berthoumieu S, Fichaux-Bourin P, et al. Dyssynchrony and perinatal psychopathology impact of child disease on parents-child interactions, the paradigm of Prader Willi syndrom. J Physiol Paris. 2016;110:427–33.
Swaab DF. Prader—Willi syndrome and the hypothalamus. Acta Paediatr. 1997;86:50–4.
Althammer F, Muscatelli F, Grinevich V, Schaaf CP. Oxytocin-based therapies for treatment of Prader-Willi and Schaaf-Yang syndromes: evidence, disappointments, and future research strategies. Transl Psychiatry. 2022;12:318.
Tauber M, Mantoulan C, Copet P, Jauregui J, Demeer G, Diene G, et al. Oxytocin may be useful to increase trust in others and decrease disruptive behaviours in patients with Prader-Willi syndrome: a randomised placebo-controlled trial in 24 patients. Orphanet J Rare Dis. 2011;6:47.
Tauber M, Boulanouar K, Diene G, Çabal-Berthoumieu S, Ehlinger V, Fichaux-Bourin P, et al. The use of oxytocin to improve feeding and social skills in infants with Prader–Willi syndrome. Pediatrics. 2017;139:e20162976.
Salles J, Lacassagne E, Eddiry S, Franchitto N, Salles JP, Tauber M. What can we learn from PWS and SNORD116 genes about the pathophysiology of addictive disorders? Mol Psychiatry. 2021;26:51–9.
Bieth E, Eddiry S, Gaston V, Lorenzini F, Buffet A, Conte Auriol F, et al. Highly restricted deletion of the SNORD116 region is implicated in Prader-Willi Syndrome. Eur J Hum Genet EJHG. 2015;23:252–5.
Huang W, Li H, Yu Q, Xiao W, Wang DO. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J Exp Clin Cancer Res. 2022;41:100.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Salles J, Eddiry S, Lacassagne E, Laurier V, Molinas C, Bieth É, et al. Patients with PWS and related syndromes display differentially methylated regions involved in neurodevelopmental and nutritional trajectory. Clin Epigenetics. 2021;13:159.
Leuner B, Caponiti JM, Gould E. Oxytocin stimulates adult neurogenesis even under conditions of stress and elevated glucocorticoids. Hippocampus. 2012;22:861–8.
Lestanova Z, Bacova Z, Kiss A, Havranek T, Strbak V, Bakos J. Oxytocin increases neurite length and expression of cytoskeletal proteins associated with neuronal growth. J Mol Neurosci. 2016;59:184–92.
Waldhorn I, Turetsky T, Steiner D, Gil Y, Benyamini H, Gropp M, et al. Modeling sex differences in humans using isogenic induced pluripotent stem cells. Stem Cell Rep. 2022;17:2732–44.
Burnett LC, LeDuc CA, Sulsona CR, Paull D, Eddiry S, Levy B, et al. Induced pluripotent stem cells (iPSC) created from skin fibroblasts of patients with Prader-Willi syndrome (PWS) retain the molecular signature of PWS. Stem Cell Res. 2016;17:526–30.
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80.
Reyes S, Fu Y, Double K, Thompson L, Kirik D, Paxinos G, et al. GIRK2 expression in dopamine neurons of the substantia nigra and ventral tegmental area. J Comp Neurol. 2012;520:2591–607.
Grow DA, Simmons DV, Gomez JA, Wanat MJ, McCarrey JR, Paladini CA, et al. Differentiation and characterization of dopaminergic neurons from baboon induced pluripotent stem cells. Stem Cells Transl Med. 2016;5:1133–44.
Hartfield EM, Yamasaki-Mann M, Fernandes HJR, Vowles J, James WS, Cowley SA, et al. Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS ONE. 2014;9:e87388.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–2.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma Oxf Engl. 2011;27:1571–2.
Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinforma Oxf Engl. 2014;30:2414–22.
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–5.
DAVID Functional annotation bioinformatics microarray analysis [Internet]. 2023. Available from: https://david.ncifcrf.gov/
Correa-da-Silva F, Fliers E, Swaab DF, Yi CX. Hypothalamic neuropeptides and neurocircuitries in Prader Willi syndrome. J Neuroendocrinol. 2021;33:e12994.
Muench C, Wiers CE, Cortes CR, Momenan R, Lohoff FW. Dopamine transporter gene methylation is associated with nucleus accumbens activation during reward processing in healthy but not alcohol-dependent individuals. Alcohol Clin Exp Res. 2018;42:21–31.
Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009;126:51–90.
Kuc K, Bielecki M, Racicka-Pawlukiewicz E, Czerwinski MB, Cybulska-Klosowicz A. The SLC6A3 gene polymorphism is related to the development of attentional functions but not to ADHD. Sci Rep. 2020;10:6176.
Schmitt KC, Rothman RB, Reith MEA. Nonclassical pharmacology of the dopamine transporter: atypical inhibitors, allosteric modulators, and partial substrates. J Pharm Exp Ther. 2013;346:2–10.
German CL, Baladi MG, McFadden LM, Hanson GR, Fleckenstein AE. Regulation of the dopamine and vesicular monoamine transporters: pharmacological targets and implications for disease. Pharm Rev. 2015;67:1005–24.
Frieling H, Römer KD, Scholz S, Mittelbach F, Wilhelm J, De Zwaan M, et al. Epigenetic dysregulation of dopaminergic genes in eating disorders. Int J Eat Disord. 2010;43:577–83.
Shumay E, Fowler JS, Volkow ND. Genomic features of the human dopamine transporter gene and its potential epigenetic states: implications for phenotypic diversity. PLoS ONE. 2010;5:e11067.
Reith MEA, Kortagere S, Wiers CE, Sun H, Kurian MA, Galli A, et al. The dopamine transporter gene SLC6A3: multidisease risks. Mol Psychiatry. 2022;27:1031–46.
Blum K, Kazmi S, Modestino EJ, Downs BW, Bagchi D, Baron D, et al. A novel precision approach to overcome the ‘addiction pandemic’ by incorporating Genetic Addiction Risk Severity (GARS) and dopamine homeostasis restoration. J Pers Med. 2021;11:212.
Heinrich H, Grunitz J, Stonawski V, Frey S, Wahl S, Albrecht B, et al. Attention, cognitive control and motivation in ADHD: linking event-related brain potentials and DNA methylation patterns in boys at early school age. Sci Rep. 2017;7:3823.
Fageera W, Chaumette B, Fortier MÈ, Grizenko N, Labbe A, Sengupta SM, et al. Association between COMT methylation and response to treatment in children with ADHD. J Psychiatr Res. 2021;135:86–93.
Dammann G, Teschler S, Haag T, Altmüller F, Tuczek F, Dammann RH. Increased DNA methylation of neuropsychiatric genes occurs in borderline personality disorder. Epigenetics. 2011;6:1454–62.
Thomas M, Banet N, Wallisch A, Glowacz K, Becker-Sadzio J, Gundel F, et al. Differential COMT DNA methylation in patients with borderline personality disorder: genotype matters. Eur Neuropsychopharmacol. 2019;29:1295–300.
Wilens TE, Martelon M, Joshi G, Bateman C, Fried R, Petty C, et al. Does ADHD predict substance use disorders? A 10-year follow-up study of young adults with ADHD. J Am Acad Child Adolesc Psychiatry. 2011;50:543–53.
Kienast T, Stoffers J, Bermpohl F, Lieb K. Borderline personality disorder and comorbid addiction. Epidemiol Treat Dtsch Arzteblatt Int 2014;111:280–6.
Hartung JE, Eskew O, Wong T, Tchivileva IE, Oladosu FA, O’Buckley SC, et al. Nuclear factor-kappa B regulates pain and COMT expression in a rodent model of inflammation. Brain Behav Immun. 2015;50:196–202.
Leriche M, Cote-Vélez A, Méndez M. Presence of pro-opiomelanocortin mRNA in the rat medial prefrontal cortex, nucleus accumbens and ventral tegmental area: studies by RT-PCR and in situ hybridization techniques. Neuropeptides. 2007;41:421–31.
Lindblom J, Opmane B, Mutulis F, Mutule I, Petrovska R, Klusa V, et al. The MC4 receptor mediates alpha-MSH induced release of nucleus accumbens dopamine. Neuroreport. 2001;12:2155–8.
Andino LM, Ryder DJ, Shapiro A, Matheny MK, Zhang Y, Judge MK, et al. POMC overexpression in the ventral tegmental area ameliorates dietary obesity. J Endocrinol. 2011;210:199–207.
Efrat S. Epigenetic memory: lessons from IPS cells derived from human β cells. Front Endocrinol. 2021;11:1063.
Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, et al. Genetic variation, not cell type of origin, underlies the majority of identifiable regulatory differences in iPSCs. PLoS Genet. 2016;12:e1005793.
Eddiry S, Diene G, Molinas C, Salles J, Auriol FC, Gennero I, et al. SNORD116 and growth hormone therapy impact IGFBP7 in Prader–Willi syndrome. Genet Med. 2021;23:1664–72.
Goetjen A, Watson M, Lieberman R, Clinton K, Kranzler HR, Covault J. Induced pluripotent stem cell reprogramming-associated methylation at the GABRA2 promoter and chr4p12 GABAA subunit gene expression in the context of alcohol use disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2020;183:464–74.
Marakulina D, Vorontsov IE, Kulakovskiy IV, Lennartsson A, Drabløs F, Medvedeva YA. EpiFactors 2022: expansion and enhancement of a curated database of human epigenetic factors and complexes. Nucleic Acids Res. 2023;51:D564–70.
Dreher JC, Kohn P, Kolachana B, Weinberger DR, Berman KF. Variation in dopamine genes influences responsivity of the human reward system. Proc Natl Acad Sci USA. 2009;106:617–22.
Congdon E, Constable RT, Lesch KP, Canli T. Influence of SLC6A3 and COMT variation on neural activation during response inhibition. Biol Psychol. 2009;81:144–52.
Hersrud SL, Stoltenberg SF. Epistatic interaction between COMT and DAT1 genes on eating behavior: a pilot study. Eat Behav. 2009;10:131–3.
Lin R, Kos A, Lopez JP, Dine J, Fiori LM, Yang J, et al. SNORD90 induces glutamatergic signaling following treatment with monoaminergic antidepressants. eLife. 2023;12:e85316.
Acknowledgements
This work was supported by a grant from the French Association for Prader-Willi Syndrome (grant R15062BB). MGX acknowledges financial support from the France Génomique National infrastructure, funded as part of “Investissement d’Avenir” program managed by the Agence Nationale pour la Recherche (contract ANR-10-INBS-09).
Author information
Authors and Affiliations
Contributions
JS, MT, and JPS developed the initial study design. NF, IG, and FJ provided feedback to improve the design. JS, SA, EL, MG and SE performed the cell culture and RT-qPCR. XA and GS performed DNA methylation and related bioinformatics analysis. EL performed RNA sequencing and related bioinformatic analysis. JS and MT performed the combined statistical analysis. JS, MT, JPS and SE wrote the initial manuscript. All authors contributed to improving the manuscript and responding to the reviewers.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salles, J., Eddiry, S., Amri, S. et al. Differential DNA methylation in iPSC-derived dopaminergic neurons: a step forward on the role of SNORD116 microdeletion in the pathophysiology of addictive behavior in Prader-Willi syndrome. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-024-02542-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-024-02542-4