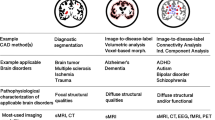
Abstract
Applications of machine learning in the biomedical sciences are growing rapidly. This growth has been spurred by diverse cross-institutional and interdisciplinary collaborations, public availability of large datasets, an increase in the accessibility of analytic routines, and the availability of powerful computing resources. With this increased access and exposure to machine learning comes a responsibility for education and a deeper understanding of its bases and bounds, borne equally by data scientists seeking to ply their analytic wares in medical research and by biomedical scientists seeking to harness such methods to glean knowledge from data. This article provides an accessible and critical review of machine learning for a biomedically informed audience, as well as its applications in psychiatry. The review covers definitions and expositions of commonly used machine learning methods, and historical trends of their use in psychiatry. We also provide a set of standards, namely Guidelines for REporting Machine Learning Investigations in Neuropsychiatry (GREMLIN), for designing and reporting studies that use machine learning as a primary data-analysis approach. Lastly, we propose the establishment of the Machine Learning in Psychiatry (MLPsych) Consortium, enumerate its objectives, and identify areas of opportunity for future applications of machine learning in biological psychiatry. This review serves as a cautiously optimistic primer on machine learning for those on the precipice as they prepare to dive into the field, either as methodological practitioners or well-informed consumers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alpaydin E. Machine learning, revised and updated edition. The MIT Press; 2021.
Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–30.
Tarca AL, Carey VJ, Chen X-W, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3:e116.
de Ridder D, de Ridder J, Reinders MJT. Pattern recognition in bioinformatics. Brief Bioinform. 2013;14:633–47.
Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–8.
Li A, Walling J, Ahn S, Kotliarov Y, Su Q, Quezado M, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69:2091–9.
Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:28–38.
Cheng W-Y, Ou Yang T-H, Anastassiou D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci Transl Med. 2013;5:181ra50.
Cheng W-Y, Ou Yang T-H, Anastassiou D. Biomolecular events in cancer revealed by attractor metagenes. PLoS Comput Biol. 2013;9:e1002920.
Gao M, Igata H, Takeuchi A, Sato K, Ikegaya Y. Machine learning-based prediction of adverse drug effects: An example of seizure-inducing compounds. J Pharm Sci. 2017;133:70–8.
Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: a review of computational problems and data sets. Proc IEEE. 2016;104:176–97.
Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. New York, NY: Springer; 2009.
Varghese B, Chen F, Hwang D, Palmer SL, De Castro Abreu AL, Ukimura O, et al. Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images. Sci Rep. 2019;9:1570.
Pandey G, Pandey OP, Rogers AJ, Ahsen ME, Hoffman GE, Raby BA, et al. A nasal brush-based classifier of asthma identified by machine learning analysis of nasal RNA sequence data. Sci Rep. 2018;8:8826.
Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat Rev Genet. 2018;19:299–310.
Zhang K, Sun Y, Wu S, Zhou M, Zhang X, Zhou R, et al. Systematic imaging in medicine: a comprehensive review. Eur J Nucl Med Mol Imaging. 2021;48:1736–58.
Oliveira AL. Biotechnology, Big Data and Artificial Intelligence. Biotechnol J. 2019;14:e1800613.
Iniesta R, Stahl D, McGuffin P. Machine learning, statistical learning and the future of biological research in psychiatry. Psychol Med. 2016;46:2455–65.
Kuhn M, Johnson K. Applied predictive modeling. New York, NY: Springer; 2013.
Lee Y, Ragguett R-M, Mansur RB, Boutilier JJ, Rosenblat JD, Trevizol A, et al. Applications of machine learning algorithms to predict therapeutic outcomes in depression: a meta-analysis and systematic review. J Affect Disord. 2018;241:519–32.
Bzdok D, Meyer-Lindenberg A. Machine learning for precision psychiatry: opportunities and challenges. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3:223–30.
Vieira S, Pinaya WHL, Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci Biobehav Rev. 2017;74:58–75.
Dwyer DB, Falkai P, Koutsouleris N. Machine learning approaches for clinical psychology and psychiatry. Annu Rev Clin Psychol. 2018;14:91–118.
Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci Biobehav Rev. 2015;57:328–49.
Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM, et al. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry. 2012;17:494–502.
Bahn S, Noll R, Barnes A, Schwarz E, Guest PC. Challenges of introducing new biomarker products for neuropsychiatric disorders into the market. Int Rev Neurobiol. 2011;101:299–327.
Lee G, Nho K, Kang B, Sohn K-A, Kim D. Predicting Alzheimer’s disease progression using multi-modal deep learning approach. Sci Rep. 2019;9:1952.
Cui R, Liu M. RNN-based longitudinal analysis for diagnosis of Alzheimer’s disease. Comput Med Imaging Graph. 2019;73:1–10.
Bellazzi R, Zupan B. Towards knowledge-based gene expression data mining. J Biomed Inf. 2007;40:787–802.
Zhang C, Zhang S. Association rule mining: models and algorithms. Berlin, Heidelberg: Springer-Verlag; 2002.
Chandola V, Banerjee A, Kumar V. Anomaly detection: a survey. ACM Comput Surv. 2009;41:15:1–15:58.
Noto K, Majidi S, Edlow AG, Wick HC, Bianchi DW, Slonim DK. CSAX: characterizing systematic anomalies in eXpression data. J Comput Biol. 2015;22:402–13.
Quinn TP, Nguyen T, Lee SC, Venkatesh S. Cancer as a tissue anomaly: classifying tumor transcriptomes based only on healthy data. Front Genet. 2019;10:599.
Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–80.
Pinaya WHL, Mechelli A, Sato JR. Using deep autoencoders to identify abnormal brain structural patterns in neuropsychiatric disorders: a large-scale multi-sample study. Hum Brain Mapp. 2019;40:944–54.
Zhang-James Y, Buitelaar JK, Rooij D, Faraone SV, The ENIGMA-ASD Working Group. Ensemble classification of autism spectrum disorder using structural magnetic resonance imaging features. JCPP Adv. 2021;1:e12042.
Geddes TA, Kim T, Nan L, Burchfield JG, Yang JYH, Tao D, et al. Autoencoder-based cluster ensembles for single-cell RNA-seq data analysis. BMC Bioinform. 2019;20:660.
Gligorijevic V, Barot M, Bonneau R. deepNF: deep network fusion for protein function prediction. Bioinformatics. 2018;34:3873–81.
Zhu X, Goldberg AB. Introduction to Semi-Supervised Learning. Synth Lect Artif Intell Mach Learn. 2009;3:1–130.
Mitchell TM. Machine learning. 1st ed. New York: McGraw-Hill Education; 1997.
Demšar J. Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res. 2006;7:1–30.
Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods. 2018;15:233–4.
Breen MS, Tylee DS, Maihofer AX, Neylan TC, Mehta D, Binder EB, et al. PTSD blood transcriptome mega-analysis: shared inflammatory pathways across biological sex and modes of trauma. Neuropsychopharmacology. 2018;43:469–81.
Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:494–502.
Vawter MP, Philibert R, Rollins B, Ruppel PL, Osborn TW. Exon array biomarkers for the differential diagnosis of schizophrenia and bipolar disorder. Mol Neuropsychiatry. 2018;3:197–213.
Nazeen S, Palmer NP, Berger B, Kohane IS. Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities. Genome Biol. 2016;17:228.
Kong SW, Collins CD, Shimizu-Motohashi Y, Holm IA, Campbell MG, Lee I-H, et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS One. 2012;7:e49475.
Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016;16:52.
Tsuang MT, Nossova N, Yager T, Tsuang M-M, Guo S-C, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5.
Tylee DS, Hess JL, Quinn TP, Barve R, Huang H, Zhang-James Y, et al. Blood transcriptomic comparison of individuals with and without autism spectrum disorder: a combined-samples mega-analysis. Am J Med Genet B Neuropsychiatr Genet. 2017;174:181–201.
Takahashi M, Hayashi H, Watanabe Y, Sawamura K, Fukui N, Watanabe J, et al. Diagnostic classification of schizophrenia by neural network analysis of blood-based gene expression signatures. Schizophr Res. 2010;119:210–8.
Zhang H, Xie Z, Yang Y, Zhao Y, Zhang B, Fang J. The correlation-base-selection algorithm for diagnostic schizophrenia based on blood-based gene expression signatures. Biomed Res Int. 2017;2017:7860506.
Yi Z, Li Z, Yu S, Yuan C, Hong W, Wang Z, et al. Blood-based gene expression profiles models for classification of subsyndromal symptomatic depression and major depressive disorder. PLoS ONE. 2012;7:e31283.
Struyf J, Dobrin S, Page D. Combining gene expression, demographic and clinical data in modeling disease: a case study of bipolar disorder and schizophrenia. BMC Genomics. 2008;9:531.
Breen MS, Uhlmann A, Nday CM, Glatt SJ, Mitt M, Metsalpu A, et al. Candidate gene networks and blood biomarkers of methamphetamine-associated psychosis: an integrative RNA-sequencing report. Transl Psychiatry. 2016;6:e802.
Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, et al. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr Res. 2016;176:114–24.
Nicodemus KK, Malley JD. Predictor correlation impacts machine learning algorithms: implications for genomic studies. Bioinformatics. 2009;25:1884–90.
Nicodemus KK, Malley JD, Strobl C, Ziegler A. The behaviour of random forest permutation-based variable importance measures under predictor correlation. BMC Bioinform. 2010;11:110.
Yassin W, Nakatani H, Zhu Y, Kojima M, Owada K, Kuwabara H, et al. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Transl Psychiatry. 2020;10:278.
Cho G, Yim J, Choi Y, Ko J, Lee S-H. Review of machine learning algorithms for diagnosing mental illness. Psychiatry Investig. 2019;16:262–9.
Zhang-James Y, Chen Q, Kuja-Halkola R, Lichtenstein P, Larsson H, Faraone SV. Machine-Learning prediction of comorbid substance use disorders in ADHD youth using Swedish registry data. J Child Psychol Psychiatry. 2020;61:1370–9.
Zhang-James Y, Razavi AS, Hoogman M, Franke B, Faraone SV. Machine learning and MRI-based diagnostic models for ADHD: are we there yet? J Atten Disord. 2023;27:335–53.
Chekroud AM, Zotti RJ, Shehzad Z, Gueorguieva R, Johnson MK, Trivedi MH, et al. Cross-trial prediction of treatment outcome in depression: a machine learning approach. Lancet Psychiatry. 2016;3:243–50.
Nie Z, Vairavan S, Narayan VA, Ye J, Li QS. Predictive modeling of treatment resistant depression using data from STAR*D and an independent clinical study. PLoS ONE. 2018;13:e0197268.
Lenhard F, Sauer S, Andersson E, Månsson KN, Mataix-Cols D, Rück C, et al. Prediction of outcome in internet-delivered cognitive behaviour therapy for paediatric obsessive-compulsive disorder: a machine learning approach. Int J Methods Psychiatr Res. 2018;27:e1576.
Flygare O, Enander J, Andersson E, Ljótsson B, Ivanov VZ, Mataix-Cols D, et al. Predictors of remission from body dysmorphic disorder after internet-delivered cognitive behavior therapy: a machine learning approach. BMC Psychiatry. 2020;20:247.
van Breda W, Bremer V, Becker D, Hoogendoorn M, Funk B, Ruwaard J, et al. Predicting therapy success for treatment as usual and blended treatment in the domain of depression. Internet Inter. 2018;12:100–4.
Wolpert DH, Macready WG. No free lunch theorems for optimization. IEEE Trans Evol Comput. 1997;1:67–82.
Koohy H. The rise and fall of machine learning methods in biomedical research. F1000Res. 2017;6:2012.
Molnar C. Interpretable machine learning. 2020. Lulu.com.
Gountouna V-E, Bermingham M, Kuznetsova K, Urda Munoz D, Agakov F, Robson S, et al. Predictive machine learning for personalised medicine in major depressive disorder. medRxiv. 2022. https://doi.org/10.1101/2022.02.11.22270724.
Strobl C, Boulesteix A-L, Zeileis A, Hothorn T. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinform. 2007;8:25.
Boulesteix A-L, Slawski M. Stability and aggregation of ranked gene lists. Brief Bioinform. 2009;10:556–68.
Verma S, Dickerson J, Hines K. Counterfactual explanations for machine learning: challenges revisited. arXiv. 2021. https://doi.org/10.48550/arXiv.2106.07756.
Lundberg S, Lee S-I. A unified approach to interpreting model predictions. arXiv. 2017. https://doi.org/10.48550/arXiv.1705.07874.
Tsang M, Liu H, Purushotham S, Murali P, Liu Y. Neural interaction transparency (NIT): disentangling learned interactions for improved interpretability. 2018:5809–18.
Zhang Y, Yang Q. A Survey on Multi-Task Learning. IEEE Trans Knowl Data Eng. 2021. 1–1
Widmer C, Rätsch G. Multitask learning in computational biology. In: Guyon I, Dror G, Lemaire V, Taylor G, Silver D, editors. Proc. ICML Workshop Unsupervised Transf. Learn., vol. 27, Bellevue, Washington, USA.
Li Y, Wang J, Ye J, Reddy CK. A multi-task learning formulation for survival analysis. KDD 16, New York, NY, USA: ACM; 2016. p. 1715–24.
Yuan H, Paskov I, Paskov H, González AJ, Leslie CS. Multitask learning improves prediction of cancer drug sensitivity. Sci Rep. 2016;6:31619.
Feriante J. Massively multitask deep learning for drug discovery. University of Wisconsin-Madison, 2015.
Xu Q, Pan SJ, Xue HH, Yang Q. Multitask learning for protein subcellular location prediction. IEEEACM Trans Comput Biol Bioinform. 2011;8:748–59.
Zhou J, Liu J, Narayan VA, Ye J, Alzheimer’s Disease Neuroimaging Initiative. Modeling disease progression via multi-task learning. Neuroimage. 2013;78:233–48.
Perlich C. Learning curves in machine learning. In: Encyclopedia of machine learning and data mining, Boston, MA: Springer US; 2011. p. 577–80.
Barnett E, Onete D, Salekin A, Faraone SV Genomic machine learning meta-regression: Insights on associations of study features with reported model performance. BioRxiv. 2022. https://doi.org/10.1101/2022.01.10.22268751.
Stripelis D, Gupta U, Saleem H, Dhinagar N, Ghai T, Sanchez R, et al. Secure federated learning for neuroimaging. arXiv. 2022. https://doi.org/10.48550/arXiv.2205.05249.
Vaid A, Jaladanki SK, Xu J, Teng S, Kumar A, Lee S, et al. Federated Learning of Electronic Health Records to Improve Mortality Prediction in Hospitalized Patients With COVID-19: Machine Learning Approach. JMIR Med Inf. 2021;9:e24207.
Zhai Y, Ong Y-S, Tsang IW. The Emerging ‘Big Dimensionality’. IEEE Comput Intell Mag. 2014;9:14–26.
Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–17.
Tang J, Alelyani S, Liu H. Feature selection for classification: a review. data classif. Algorithms Appl., CRC Press; 2014. p. 37–64.
Sorzano COS, Vargas J, Pascual Montano A. A survey of dimensionality reduction techniques. ArXiv. 2014. https://doi.org/10.48550/arXiv.1403.2877
Smialowski P, Frishman D, Kramer S. Pitfalls of supervised feature selection. Bioinformatics. 2010;26:440–3.
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A. Feature selection for high-dimensional data. Springer; 2015.
Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci USA. 2002;99:6562–6.
Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
Quinn TP, Lee SC, Venkatesh S, Nguyen T. Improving the classification of neuropsychiatric conditions using gene ontology terms as features. Am J Med Genet B Neuropsychiatr Genet. 2019;180:508–18.
Chawla NV Data Mining for Imbalanced Datasets: An Overview. In: Maimon O, Rokach L, editors. Data mining and knowledge discovery handbook. Boston, MA: Springer US; 2005. p. 853–67.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10:e0118432.
Altman N, Krzywinski M. Graphical assessment of tests and classifiers. Nat Methods. 2021;18:840–2.
Zhao Q, Adeli E, Pohl KM. Training confounder-free deep learning models for medical applications. Nat Commun. 2020;11:6010.
Loughman A, Quinn T, Nation ML, Reichelt A, Moore RJ, Van TTH, et al. Infant microbiota in colic: predictive associations with problem crying and subsequent child behavior. J Dev Orig Health Dis. 2021;12:260–70.
Wang H, Wu Z, Xing EP. Removing confounding factors associated weights in deep neural networks improves the prediction accuracy for healthcare applications. Pac Symp Biocomput. 2019;24:54–65.
Ganin Y, Ustinova E, Ajakan H, Germain P, Larochelle H, Laviolette F, et al. Domain-adversarial training of neural networks. J Mach Learn Res. 2016;17:2096–30.
Liu Y, Nyunoya T, Leng S, Belinsky SA, Tesfaigzi Y, Bruse S. Softwares and methods for estimating genetic ancestry in human populations. Hum Genomics. 2013;7:1.
Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35.
Rajkomar A, Hardt M, Howell MD, Corrado G, Chin MH. Ensuring Fairness in Machine Learning to Advance Health Equity. Ann Intern Med. 2018;169:866–72.
Quinn TP, Coghlan S. Readying medical students for medical AI: the need to embed AI ethics education. ArXiv. 2021. https://doi.org/10.48550/arXiv.2109.02866.
Food and Drug Administration (FDA). Artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD) action plan.
Franco D, Oneto L, Navarin N, Anguita D. Toward learning trustworthily from data combining privacy, fairness, and explainability: an application to face recognition. Entropy Basel Switz. 2021;23:1047.
Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, et al. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet. 2010;127:441–52.
Nicodemus KK, Law AJ, Radulescu E, Luna A, Kolachana B, Vakkalanka R, et al. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Arch Gen Psychiatry. 2010;67:991–1001.
Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22.
Song X, Liu X, Liu F, Wang C. Comparison of machine learning and logistic regression models in predicting acute kidney injury: A systematic review and meta-analysis. Int J Med Inf. 2021;151:104484.
Smith DL, Held P. Moving toward precision PTSD treatment: predicting veterans’ intensive PTSD treatment response using continuously updating machine learning models. Psychol Med. 2023;53:5500–9.
Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, et al. Transcriptomic abnormalities in peripheral blood in bipolar disorder, and discrimination of the major psychoses. Schizophr Res. 2020;217:124–35.
Zitnik M, Nguyen F, Wang B, Leskovec J, Goldenberg A, Hoffman MM. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf Fusion. 2019;50:71–91.
Schwarz E, Leweke FM, Bahn S, Liò P. Clinical bioinformatics for complex disorders: a schizophrenia case study. BMC Bioinform. 2009;10:S6.
Xia CH, Ma Z, Ciric R, Gu S, Betzel RF, Kaczkurkin AN, et al. Linked dimensions of psychopathology and connectivity in functional brain networks. Nat Commun. 2018;9:3003.
Shomorony I, Cirulli ET, Huang L, Napier LA, Heister RR, Hicks M, et al. Unsupervised integration of multimodal dataset identifies novel signatures of health and disease. BioRxiv. 2018. https://doi.org/10.1101/432641.
Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, et al. Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 2018;14:e8124.
Koutsouleris N, Meisenzahl EM, Borgwardt S, Riecher-Rössler A, Frodl T, Kambeitz J, et al. Individualized differential diagnosis of schizophrenia and mood disorders using neuroanatomical biomarkers. Brain. 2015;138:2059–73.
Doan NT, Kaufmann T, Bettella F, Jørgensen KN, Brandt CL, Moberget T, et al. Distinct multivariate brain morphological patterns and their added predictive value with cognitive and polygenic risk scores in mental disorders. Neuroimage Clin. 2017;15:719–31.
Cao H, Duan J, Lin D, Shugart YY, Calhoun V, Wang Y-P. Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs. Neuroimage. 2014;102:220–8.
Li YC, Wang L, Law JN, Murali TM, Pandey G. Integrating multimodal data through interpretable heterogeneous ensembles. Bioinforma Adv. 2022;2:vbac065.
Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15:20170387.
Lin E, Kuo P-H, Liu Y-L, Yu YW-Y, Yang AC, Tsai S-J. A deep learning approach for predicting antidepressant response in major depression using clinical and genetic biomarkers. Front Psychiatry. 2018;9:290.
Sundaram L, Bhat RR, Viswanath V, Li X. DeepBipolar: Identifying genomic mutations for bipolar disorder via deep learning. Hum Mutat. 2017;38:1217–24.
Plis SM, Hjelm DR, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, et al. Deep learning for neuroimaging: a validation study. Front Neurosci. 2014;8:229.
Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–92.
Bulik-Sullivan, Loh BK, Finucane P-R, Ripke HK, Yang S, Schizophrenia Working Group of the Psychiatric Genomics Consortium J, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Krapohl E, Patel H, Newhouse S, Curtis CJ, von Stumm S, Dale PS, et al. Multi-polygenic score approach to trait prediction. Mol Psychiatry. 2018;23:1368–74.
Barnett EJ, Biederman J, Doyle AE, Hess J, DiSalvo M, Faraone SV. Identifying pediatric mood disorders from transdiagnostic polygenic risk scores: a study of children and adolescents. J Clin Psychiatry. 2022;83:40635.
Chen J, Schwarz E. BioMM: Biologically-informed Multi-stage Machine learning for identification of epigenetic fingerprints. arXiv:171200336. 2017. https://doi.org/10.48550/arXiv.1712.00336.
Vu M-AT, Adalı T, Ba D, Buzsáki G, Carlson D, Heller K, et al. A shared vision for machine learning in neuroscience. J Neurosci. 2018;38:1601–7.
Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2:230–43.
Kang J, Rancati T, Lee S, Oh JH, Kerns SL, Scott JG, et al. Machine learning and radiogenomics: lessons learned and future directions. Front Oncol. 2018;8:228.
Shrager J, Tenenbaum JM. Rapid learning for precision oncology. Nat Rev Clin Oncol. 2014;11:109.
Doyle-Lindrud S. Watson will see you now: a supercomputer to help clinicians make informed treatment decisions. Clin J Oncol Nurs. 2015;19:31–2.
Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16:933–51.
Neale B. Perspective on data sharing and open science in psychiatric genetics. Eur Neuropsychopharmacol. 2019;29:S778.
Bell V. Open science in mental health research. Lancet Psychiatry. 2017;4:525–6.
Hardwicke TE, Mathur MB, MacDonald K, Nilsonne G, Banks GC, Kidwell MC, et al. Data availability, reusability, and analytic reproducibility: evaluating the impact of a mandatory open data policy at the journal Cognition. R Soc Open Sci. 2018;5:180448.
National Research Council (US) Committee on Applied, Statistics T. The current state of data integration in science. USA: National Academies Press; 2010.
Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry. 2018;175:15–27.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–D901.
Shen Z, Spruit M. A systematic review of open source clinical software on GitHub for improving software reuse in smart healthcare. NATO Adv Sci Inst Ser E Appl Sci. 2019;9:150.
Guinney J, Saez-Rodriguez J. Alternative models for sharing confidential biomedical data. Nat Biotechnol. 2018;36:391–2.
Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84.
Lapatas V, Stefanidakis M, Jimenez RC, Via A, Schneider MV. Data integration in biological research: an overview. J Biol Res. 2015;22:9.
Lam RW, Milev R, Rotzinger S, Andreazza AC, Blier P, Brenner C, et al. Discovering biomarkers for antidepressant response: protocol from the Canadian biomarker integration network in depression (CAN-BIND) and clinical characteristics of the first patient cohort. BMC Psychiatry. 2016;16:105.
Blum AL, Langley P. Selection of relevant features and examples in machine learning. Artif Intell. 1997;97:245–71.
Kolachalama VB, Garg PS. Machine learning and medical education. Npj Digit Med. 2018;1:54.
Hekler A, Utikal JS, Enk AH, Solass W, Schmitt M, Klode J, et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur J Cancer Oxf Engl. 2019;118:91–6.
McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577:89–94.
Wang S, Manning C. Fast dropout training. In: Dasgupta S, McAllester D, editors. Proc. 30th International Conference on Machine Learning, vol. 28, PMLR: Atlanta, Georgia, USA; 2013. p. 118–26.
Lakshminarayanan B, Pritzel A, Blundell C. Simple and scalable predictive uncertainty estimation using deep ensembles. ArXiv. 2017. https://doi.org/10.48550/arXiv.1612.01474.
Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing. 2019;338:34–45.
Dolezal JM, Srisuwananukorn A, Karpeyev D, Ramesh S, Kochanny S, Cody B, et al. Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology. Nat Commun. 2022;13:6572.
Tandon N, Tandon R. Machine learning in psychiatry- standards and guidelines. Asian J Psychiatr. 2019;44:A1–A4.
Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. ArXiv. 2019. https://doi.org/10.48550/arXiv.1910.10045.
Bollen KA, Jackman RW. Regression diagnostics: an expository treatment of outliers and influential cases. Socio Methods Res. 1985;13:510–42.
Samiei M, Würfl T, Deleu T, Weiss M, Dutil F, Fevens T, et al. The TCGA meta-dataset clinical benchmark. ArXiv. 2019. https://doi.org/10.48550/arXiv.1910.08636.
Feng J, Xu H, Mannor S, Yan S. Robust logistic regression and classification. In: Advances in neural information processing systems, vol. 27, Curran Associates, Inc.; 2014.
Ying X. An overview of overfitting and its solutions. J Phys Confer Ser. 2019;1168:022022.
Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev. 2012;36:1140–52.
Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS ONE. 2019;14:e0224365.
Ramteke RJ, Khachane M. Automatic medical image classification and abnormality detection using K- nearest neighbour. Int J Adv Comput Res. 2012;2:190–6.
Awoyemi JO, Adetunmbi AO, Oluwadare SA. Credit card fraud detection using machine learning techniques: a comparative analysis. 2017 Int. Confer. Comput. Netw. Inform. Lagos, Nigeria: ICCNI; 2017. p. 1–9.
Delgadillo J, Lutz W. A development pathway towards precision mental health care. JAMA Psychiatry. 2020;77:889.
Lutz W, Schwartz B, Martín Gómez Penedo J, Boyle K, Deisenhofer A-K. Working towards the development and implementation of precision mental healthcare: an example. Adm Policy Ment Health. 2020;47:856–61.
Heckerman D A Bayesian Approach to Learning Causal Networks. ArXiv. 2015. https://doi.org/10.48550/arXiv.1302.4958.
Madar IH, Sultan G, Tayubi IA, Hasan AN, Pahi B, Rai A, et al. Identification of marker genes in Alzheimer’s disease using a machine-learning model. Bioinformation. 2021;17:348–55.
Chen Y-C, Wheeler TA, Kochenderfer MJ. Learning discrete bayesian networks from continuous data. J Artif Intell Res. 2017;59:103–32.
Cheng J, Liu H-P, Lin W-Y, Tsai F-J. Machine learning compensates fold-change method and highlights oxidative phosphorylation in the brain transcriptome of Alzheimer’s disease. Sci Rep. 2021;11:13704.
Dwyer K, Holte R. Decision Tree Instability and Active Learning. In: Kok JN, Koronacki J, de Mantaras RL, Matwin S, Mladenič D, Skowron A, editors. Mach. Learn. ECML 2007, Berlin, Heidelberg: Springer; 2007. p. 128–39.
Breiman L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat Sci. 2001;16:199–231.
Ainscough BJ, Barnell EK, Ronning P, Campbell KM, Wagner AH, Fehniger TA, et al. A deep learning approach to automate refinement of somatic variant calling from cancer sequencing data. Nat Genet. 2018;50:1735–43.
Sole X, Ramisa A, Torras C. Evaluation of random forests on large-scale classification problems using a Bag-of-Visual-Words representation.
Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd ed. New York, NY: Springer; 2009.
Vangay P, Hillmann BM, Knights D. Microbiome Learning Repo (ML Repo): a public repository of microbiome regression and classification tasks. GigaScience. 2019;8:giz042.
Zhang H, Nettleton D, Zhu Z. Regression-enhanced random forests. ArXiv. 2019. https://doi.org/10.48550/arXiv.1904.10416.
Hinton G, Deng L, Yu D, Dahl GE, Mohamed A, Jaitly N, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97.
Suk H-I, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013;16:583–90.
Telenti A, Lippert C, Chang P-C, DePristo M. Deep learning of genomic variation and regulatory network data. Hum Mol Genet. 2018;27:R63–R71.
Wang F, Jiang M, Qian C, Yang S, Li C, Zhang H, et al. Residual Attention Network for Image Classification. arXiv:170406904. 2017. https://doi.org/10.48550/arXiv.1704.06904.
Young T, Hazarika D, Poria S, Cambria E. Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag. 2018;13:55–75.
Zrimec J, Börlin CS, Buric F, Muhammad AS, Chen R, Siewers V, et al. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure. Nat Commun. 2020;11:6141.
Pandey M, Fernandez M, Gentile F, Isayev O, Tropsha A, Stern AC, et al. The transformational role of GPU computing and deep learning in drug discovery. Nat Mach Intell. 2022;4:211–21.
Koppe G, Meyer-Lindenberg A, Durstewitz D. Deep learning for small and big data in psychiatry. Neuropsychopharmacology. 2021;46:176–90.
Alwosheel A, van Cranenburgh S, Chorus CG. Is your dataset big enough? Sample size requirements when using artificial neural networks for discrete choice analysis. J Choice Model. 2018;28:167–82.
Passafaro TL, Lopes FB, Dórea JRR, Craven M, Breen V, Hawken RJ, et al. Would large dataset sample size unveil the potential of deep neural networks for improved genome-enabled prediction of complex traits? The case for body weight in broilers. BMC Genomics. 2020;21:771.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Acknowledgements
SJG is supported by grants from the U.S. National Institutes of Health (R01AG064955 and R21MH126494), the U.S. National Science Foundation, the Sidney R. Baer, Jr. Foundation, and NARSAD: The Brain & Behavior Research Foundation. GP is supported by a grant from the U.S. National Institutes of Health (R01HG011407) and an IBM Faculty Award. PL is supported a grant from the European Union’s Horizon 2020 research and innovation programme (634880), EDC-MixRisk, and Patient-Centered Outcome Research Institute (PCORI). JLH is supported by a grant from the U.S. National Institutes of Health (R21MH126494 and R01NS128535) and NARSAD: The Brain & Behavior Research Foundation. SVF is supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 965381; NIMH grants U01AR076092-01A1, 1R21MH1264940, R01MH116037; 1R01NS128535 – 01; Oregon Health and Science University, Otsuka Pharmaceuticals, Noven Pharmaceuticals Incorporated, and Supernus Pharmaceutical Company.
Author information
Authors and Affiliations
Consortia
Contributions
TPQ, JLH, SVF, DJM, KKN, ES, YZJ, and SJG conceived of the study or a section of the study. MSB, VSM, XM, SSME, MM, ES, A-CH, YJT, and AH performed the systematic review and data extraction. TPQ, JLH, JC, GP, ES, AS, IB, P-IL, YZJ, MEA, DJM contributed a section to the initial draft of the manuscript. VSM, MMB, AH, MM, SSME, XM, ES, YJT, MSB, EJB, YZJ, MEA, HC, JH, AS, P-IL, KKN, AML, IB, SVF, MJC, GP, DJM, and SJG critically revised the manuscript. DJM and SJG supervised the project. All authors approved the final version of the manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
SVF in the past year, received income, potential income, travel expenses continuing education support and/or research support from Aardvark, Aardwolf, AIMH, Tris, Otsuka, Ironshore, Kanjo, Johnson & Johnson/Kenvue, KemPharm/Corium, Akili, Supernus, Atentiv, Noven, Sky Therapeutics, Axsome and Genomind. With his institution, he has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. He also receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts and Elsevier: ADHD: Non-Pharmacologic Interventions. He is Program Director of www.ADHDEvidence.org and www.ADHDinAdults.com.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Quinn, T.P., Hess, J.L., Marshe, V.S. et al. A primer on the use of machine learning to distil knowledge from data in biological psychiatry. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-023-02334-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-023-02334-2