Abstract
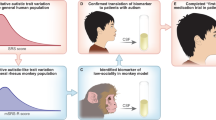
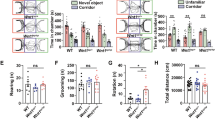
Despite intensive studies in modeling neuropsychiatric disorders especially autism spectrum disorder (ASD) in animals, many challenges remain. Genetic mutant mice have contributed substantially to the current understanding of the molecular and neural circuit mechanisms underlying ASD. However, the translational value of ASD mouse models in preclinical studies is limited to certain aspects of the disease due to the apparent differences in brain and behavior between rodents and humans. Non-human primates have been used to model ASD in recent years. However, a low reproduction rate due to a long reproductive cycle and a single birth per pregnancy, and an extremely high cost prohibit a wide use of them in preclinical studies. Canine model is an appealing alternative because of its complex and effective dog–human social interactions. In contrast to non-human primates, dog has comparable drug metabolism as humans and a high reproduction rate. In this study, we aimed to model ASD in experimental dogs by manipulating the Shank3 gene as SHANK3 mutations are one of most replicated genetic defects identified from ASD patients. Using CRISPR/Cas9 gene editing, we successfully generated and characterized multiple lines of Beagle Shank3 (bShank3) mutants that have been propagated for a few generations. We developed and validated a battery of behavioral assays that can be used in controlled experimental setting for mutant dogs. bShank3 mutants exhibited distinct and robust social behavior deficits including social withdrawal and reduced social interactions with humans, and heightened anxiety in different experimental settings (n = 27 for wild-type controls and n = 44 for mutants). We demonstrate the feasibility of producing a large number of mutant animals in a reasonable time frame. The robust and unique behavioral findings support the validity and value of a canine model to investigate the pathophysiology and develop treatments for ASD and potentially other psychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw whole-genome sequence data from founder mutants and WT parents are deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation, Chinese Academy of Sciences, under accession number CRA004090 and publicly accessible at https://bigd.big.ac.cn/gsa.
References
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84.e523.
Fu JM, Satterstrom FK, Peng M, Brand H, Collins RL, Dong S, et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat Genet. 2022;54:1320–31.
Zhou X, Feliciano P, Shu C, Wang T, Astrovskaya I, Hall JB, et al. Integrating de novo and inherited variants in 42,607 autism cases identifies mutations in new moderate-risk genes. Nat Genet. 2022;54:1305–19.
Jennings CG, Landman R, Zhou Y, Sharma J, Hyman J, Movshon JA, et al. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci. 2016;19:1123–30.
Berry-Kravis EM, Lindemann L, Jonch AE, Apostol G, Bear MF, Carpenter RL, et al. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov. 2018;17:280–99.
Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. 2016;530:98–102.
Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, et al. Modeling Rett syndrome using TALEN-Edited MECP2 mutant cynomolgus monkeys. Cell. 2017;169:945–55.e910.
Zhao H, Tu Z, Xu H, Yan S, Yan H, Zheng Y, et al. Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res. 2017;27:1293–7.
Tu Z, Zhao H, Li B, Yan S, Wang L, Tang Y, et al. CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum Mol Genet. 2019;28:561–71.
Zhou Y, Sharma J, Ke Q, Landman R, Yuan J, Chen H, et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature. 2019;570:326–31.
Zhao H, Jiang YH, Zhang YQ. Modeling autism in non-human primates: opportunities and challenges. Autism Res. 2018;11:686–94.
Feng G, Jensen FE, Greely HT, Okano H, Treue S, Roberts AC, et al. Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proc Natl Acad Sci USA. 2020;117:24022–31.
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19.
Hong H, Zhao Z, Huang X, Guo C, Zhao H, Wang GD, et al. Comparative proteome and cis-regulatory element analysis reveals specific molecular pathways conserved in dog and human brains. Mol Cell Proteom. 2022;21:100261.
Hare B, Brown M, Williamson C, Tomasello M. The domestication of social cognition in dogs. Science. 2002;298:1634–6.
Miklosi A, Kubinyi E, Topal J, Gacsi M, Viranyi Z, Csanyi V. A simple reason for a big difference: wolves do not look back at humans, but dogs do. Curr Biol. 2003;13:763–6.
Teglas E, Gergely A, Kupan K, Miklosi A, Topal J. Dogs’ gaze following is tuned to human communicative signals. Curr Biol. 2012;22:209–12.
Nagasawa M, Mitsui S, En S, Ohtani N, Ohta M, Sakuma Y, et al. Social evolution. Oxytocin-gaze positive loop and the coevolution of human-dog bonds. Science. 2015;348:333–6.
Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27.
Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–7.
Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–108.
Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat Commun. 2016;7:11459.
Anderson AC. Beagle as an experimental dog. Ames: Iowa University Press; 1970. p. 616.
Bolman B. Dogs for life: Beagles, drugs, and capital in the twentieth century. J Hist Biol. 2022;55:147–79.
Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Mol Autism. 2014;5:30.
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–43.
Siwak CT, Tapp PD, Milgram NW. Effect of age and level of cognitive function on spontaneous and exploratory behaviors in the beagle dog. Learn Mem. 2001;8:317–25.
Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302.
Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502.
Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42.
Serpell J. The domestic dog: its evolution, behaviour and interactions with people. Cambridge: Cambridge University Press; 1995.
Quaranta A, Siniscalchi M, Vallortigara G. Asymmetric tail-wagging responses by dogs to different emotive stimuli. Curr Biol. 2007;17:R199–201.
Bradshaw J, Rooney N. Dog social behavior and communication. In: Serpell J, editor. The domestic dogs. Cambridge: Cambridge University Press; 2016. p. 133–59.
Ren W, Wei P, Yu S, Zhang YQ. Left-right asymmetry and attractor-like dynamics of dog’s tail wagging during dog-human interactions. iScience. 2022;25:104747.
Siniscalchi M, d’Ingeo S, Minunno M, Quaranta A. Communication in dogs. Animals. 2018;8:131.
Muller CA, Schmitt K, Barber AL, Huber L. Dogs can discriminate emotional expressions of human faces. Curr Biol. 2015;25:601–5.
Hekman JP, Karas AZ, Sharp CR. Psychogenic stress in hospitalized dogs: cross species comparisons, implications for health care, and the challenges of evaluation. Animals. 2014;4:331–47.
Beerda B. Behavioral and hormonal indicators of enduring environmental stress in dogs. Anim Welf. 2000;2000:49–62.
Marshall-Pescini S, Colombo E, Passalacqua C, Merola I, Prato-Previde E. Gaze alternation in dogs and toddlers in an unsolvable task: evidence of an audience effect. Anim Cogn. 2013;16:933–43.
Soorya L, Kolevzon A, Zweifach J, Lim T, Dobry Y, Schwartz L, et al. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol Autism. 2013;4:18.
Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580.
De Rubeis S, Siper PM, Durkin A, Weissman J, Muratet F, Halpern D, et al. Delineation of the genetic and clinical spectrum of Phelan-McDermid syndrome caused by SHANK3 point mutations. Mol Autism. 2018;9:31.
Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15.
Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–13.
Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM. Altered striatal synaptic function and abnormal behaviour in Shank3 exon4-9 deletion mouse model of autism. Autism Res. 2016;9:350–75.
Qin L, Ma K, Wang ZJ, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–75.
Delling JP, Boeckers TM. Comparison of SHANK3 deficiency in animal models: phenotypes, treatment strategies, and translational implications. J Neurodev Disord. 2021;13:55.
Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18:147–57.
Dawson G, Lewy A. Arousal, attention, and the sociemotional impairments of individuals with autism. In: Dawson G, editor. Autism: nature, diagnosis, and treatment. New York: Guilford Press; 1989, p. 49–74.
Weiss JA, Thomson K, Chan L. A systematic literature review of emotion regulation measurement in individuals with autism spectrum disorder. Autism Res. 2014;7:629–48.
Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910.
Huang K, Han Y, Chen K, Pan H, Zhao G, Yi W, et al. A hierarchical 3D-motion learning framework for animal spontaneous behavior mapping. Nat Commun. 2021;12:2784.
Marshall JD, Aldarondo DE, Dunn TW, Wang WL, Berman GJ, Olveczky BP. Continuous whole-body 3d kinematic recordings across the rodent behavioral repertoire. Neuron. 2021;109:420–37.e428.
Ren W, Huang K, Li Y, Yang Q, Wang L, Guo K, et al. Altered pupil responses to social and non-social stimuli in Shank3 mutant dogs. Mol Psychiatry. Accepted.
Feng C, Wang X, Shi H, Yan Q, Zheng M, Li J, et al. Generation of ApoE deficient dogs via combination of embryo injection of CRISPR/Cas9 with somatic cell nuclear transfer. J Genet Genom. 2018;45:47–50.
Luo X, He Y, Zhang C, He X, Yan L, Li M, et al. Trio deep-sequencing does not reveal unexpected off-target and on-target mutations in Cas9-edited rhesus monkeys. Nat Commun. 2019;10:5525.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Acknowledgements
We thank Prof. X. Yu, X. Li, X. Xu, K. Guo, R. Zhang and A. Andics for discussion. This work was supported in part by the Ministry of Science and Technology of China (2019YFA0707100 and 2021ZD0203901), the National Natural Science Foundation of China (31830036, 31730039, and 31921002), the National Major Scientific Instruments and Equipment Development Project (ZDYZ2015-2), the Chinese Academy of Sciences Strategic Priority Research Program B grants (XDBS1020100, and XDB32010300), and Spring City Plan: the High-level Talent Promotion and Training Project of Kunming (2022SCP001).
Author information
Authors and Affiliations
Contributions
YHJ and YQZ conceptualized the project, supervised data collection and analysis. XW and LL performed gene targeting. HL, YZ, and GW analyzed sequencing data. HX and HZ performed Western analysis. RT, HZ, YL, and QS designed and performed behavioral experiments. JZ, ZS, JM, WL, and WR assisted data collection and analysis. RT, HZ, LY and YQZ wrote the manuscript, and YHJ and YQZ finalized the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, R., Li, Y., Zhao, H. et al. Modeling SHANK3-associated autism spectrum disorder in Beagle dogs via CRISPR/Cas9 gene editing. Mol Psychiatry 28, 3739–3750 (2023). https://doi.org/10.1038/s41380-023-02276-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02276-9