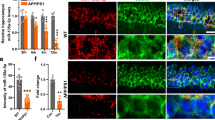
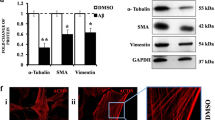
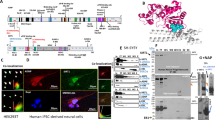
Abstract
De novo heterozygous mutations in activity-dependent neuroprotective protein (ADNP) cause autistic ADNP syndrome. ADNP mutations impair microtubule (MT) function, essential for synaptic activity. The ADNP MT-associating fragment NAPVSIPQ (called NAP) contains an MT end-binding protein interacting domain, SxIP (mimicking the active-peptide, SKIP). We hypothesized that not all ADNP mutations are similarly deleterious and that the NAPV portion of NAPVSIPQ is biologically active. Using the eukaryotic linear motif (ELM) resource, we identified a Src homology 3 (SH3) domain-ligand association site in NAP responsible for controlling signaling pathways regulating the cytoskeleton, namely NAPVSIP. Altogether, we mapped multiple SH3-binding sites in ADNP. Comparisons of the effects of ADNP mutations p.Glu830synfs*83, p.Lys408Valfs*31, p.Ser404* on MT dynamics and Tau interactions (live-cell fluorescence-microscopy) suggested spared toxic function in p.Lys408Valfs*31, with a regained SH3-binding motif due to the frameshift insertion. Site-directed-mutagenesis, abolishing the p.Lys408Valfs*31 SH3-binding motif, produced MT toxicity. NAP normalized MT activities in the face of all ADNP mutations, although, SKIP, missing the SH3-binding motif, showed reduced efficacy in terms of MT-Tau interactions, as compared with NAP. Lastly, SH3 and multiple ankyrin repeat domains protein 3 (SHANK3), a major autism gene product, interact with the cytoskeleton through an actin-binding motif to modify behavior. Similarly, ELM analysis identified an actin-binding site on ADNP, suggesting direct SH3 and indirect SHANK3/ADNP associations. Actin co-immunoprecipitations from mouse brain extracts showed NAP-mediated normalization of Shank3-Adnp-actin interactions. Furthermore, NAP treatment ameliorated aberrant behavior in mice homozygous for the Shank3 ASD-linked InsG3680 mutation, revealing a fundamental shared mechanism between ADNP and SHANK3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pinhasov A, Mandel S, Torchinsky A, Giladi E, Pittel Z, Goldsweig AM, et al. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res Dev Brain Res. 2003;144:83–90.
Mandel S, Rechavi G, Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev Biol. 2007;303:814–24.
Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E, et al. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry. 2015;20:126–32.
Ivashko-Pachima Y, Hadar A, Grigg I, Korenková V, Kapitansky O, Karmon G, et al. Discovery of autism/intellectual disability somatic mutations in Alzheimer’s brains: mutated ADNP cytoskeletal impairments and repair as a case study. Mol Psychiatry. 2021;26:1619–33.
Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–4.
Van Dijck A, Vulto-van Silfhout AT, Cappuyns E, van der Werf IM, Mancini GM, Tzschach A, et al. Clinical presentation of a complex neurodevelopmental disorder caused by mutations in ADNP. Biol Psychiatry. 2019;85:287–97.
Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–84.e23.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Mandel S, Gozes I. Activity-dependent neuroprotective protein constitutes a novel element in the SWI/SNF chromatin remodeling complex. J Biol Chem. 2007;282:34448–56.
Ostapcuk V, Mohn F, Carl SH, Basters A, Hess D, Iesmantavicius V, et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–43.
Amram N, Hacohen-Kleiman G, Sragovich S, Malishkevich A, Katz J, Touloumi O, et al. Sexual divergence in microtubule function: the novel intranasal microtubule targeting SKIP normalizes axonal transport and enhances memory. Mol Psychiatry. 2016;21:1467–76.
Gozes I, Van Dijck A, Hacohen-Kleiman G, Grigg I, Karmon G, Giladi E, et al. Premature primary tooth eruption in cognitive/motor-delayed ADNP-mutated children. Transl Psychiatry. 2017;7:e1166.
Grigg I, Ivashko-Pachima Y, Hait TA, Korenková V, Touloumi O, Lagoudaki R, et al. Tauopathy in the young autistic brain: novel biomarker and therapeutic target. Transl Psychiatry. 2020;10:228.
Sun X, Peng X, Cao Y, Zhou Y, Sun Y. ADNP promotes neural differentiation by modulating Wnt/beta-catenin signaling. Nat Commun. 2020;11:2984.
Yan Q, Wulfridge P, Doherty J, Fernandez-Luna JL, Real PJ, Tang HY, et al. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat Commun. 2022;13:53.
Ferrari R, de Llobet Cucalon LI, Di Vona C, Le Dilly F, Vidal E, Lioutas A, et al. TFIIIC binding to Alu elements controls gene expression via chromatin looping and histone acetylation. Mol Cell. 2020;77:475–87.e11.
Hadar A, Kapitansky O, Ganaiem M, Sragovich S, Lobyntseva A, Giladi E, et al. Introducing ADNP and SIRT1 as new partners regulating microtubules and histone methylation. Mol Psychiatry. 2021;26:6550–61.
Markenscoff-Papadimitriou E, Binyameen F, Whalen S, Price J, Lim K, Ypsilanti AR, et al. Autism risk gene POGZ promotes chromatin accessibility and expression of clustered synaptic genes. Cell Rep. 2021;37:110089.
Oz S, Kapitansky O, Ivashco-Pachima Y, Malishkevich A, Giladi E, Skalka N, et al. The NAP motif of activity-dependent neuroprotective protein (ADNP) regulates dendritic spines through microtubule end binding proteins. Mol Psychiatry. 2014;19:1115–24.
Ivashko-Pachima Y, Sayas CL, Malishkevich A, Gozes I. ADNP/NAP dramatically increase microtubule end-binding protein-Tau interaction: a novel avenue for protection against tauopathy. Mol Psychiatry. 2017;22:1335–44.
Vulih-Shultzman I, Pinhasov A, Mandel S, Grigoriadis N, Touloumi O, Pittel Z, et al. Activity-dependent neuroprotective protein snippet NAP reduces tau hyperphosphorylation and enhances learning in a novel transgenic mouse model. J Pharmacol Exp Ther. 2007;323:438–49.
Oz S, Ivashko-Pachima Y, Gozes I. The ADNP derived peptide, NAP modulates the tubulin pool: implication for neurotrophic and neuroprotective activities. PloS ONE. 2012;7:e51458.
Smith-Swintosky VL, Gozes I, Brenneman DE, D’Andrea MR, Plata-Salaman CR. Activity-dependent neurotrophic factor-9 and NAP promote neurite outgrowth in rat hippocampal and cortical cultures. J Mol Neurosci. 2005;25:225–38.
Hacohen-Kleiman G, Sragovich S, Karmon G, Gao AYL, Grigg I, Pasmanik-Chor M, et al. Activity-dependent neuroprotective protein deficiency models synaptic and developmental phenotypes of autism-like syndrome. J Clin Investig. 2018;128:4956–69.
Lasser M, Tiber J, Lowery LA. The role of the microtubule cytoskeleton in neurodevelopmental disorders. Front Cell Neurosci. 2018;12:165.
Karmon G, Sragovich S, Hacohen-Kleiman G, Ben-Horin-Hazak I, Kasparek P, Schuster B, et al. Novel ADNP syndrome mice reveal dramatic sex-specific peripheral gene expression with brain synaptic and Tau pathologies. Biol Psychiatry. 2021:S0006-3223(21)01630-9. https://doi.org/10.1016/j.biopsych.2021.09.018. Online ahead of print.
Morimoto BH, Schmechel D, Hirman J, Blackwell A, Keith J, Gold M, et al. A double-blind, placebo-controlled, ascending-dose, randomized study to evaluate the safety, tolerability and effects on cognition of AL-108 after 12 weeks of intranasal administration in subjects with mild cognitive impairment. Dement Geriatr Cogn Disord. 2013;35:325–36.
Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, et al. Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron. 2016;89:147–62.
Schlessinger J. SH2/SH3 signaling proteins. Curr Opin Genet Dev. 1994;4:25–30.
Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17:71–84.
Salomaa SI, Miihkinen M, Kremneva E, Paatero I, Lilja J, Jacquemet G, et al. SHANK3 conformation regulates direct actin binding and crosstalk with Rap1 signaling. Curr Biol. 2021;31:4956–70.e59.
Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18:147–57.
Barak B, Feng G. Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat Neurosci. 2016;19:647–55.
Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–7.
Ivashko-Pachima Y, Gozes I. A novel microtubule-Tau association enhancer and neuroprotective drug candidate: Ac-SKIP. Front Cell Neurosci. 2019;13:435.
Ivashko-Pachima Y, Maor-Nof M, Gozes I. NAP (davunetide) preferential interaction with dynamic 3-repeat Tau explains differential protection in selected tauopathies. PloS ONE. 2019;14:e0213666.
Sragovich S, Ziv Y, Vaisvaser S, Shomron N, Hendler T, Gozes I. The autism-mutated ADNP plays a key role in stress response. Transl Psychiatry. 2019;9:235.
Jaljuli I, Kafkafi N, Giladi E, Golani I, Gozes I, Chesler E, et al. Improving replicability using interaction with laboratories: a multi-lab experimental assessment. BioRXiv. 2021. https://doi.org/10.1101/2021.12.05.471264.
Dinkel H, Van Roey K, Michael S, Kumar M, Uyar B, Altenberg B, et al. ELM 2016-data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44:D294–300.
Reynolds CH, Garwood CJ, Wray S, Price C, Kellie S, Perera T, et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J Biol Chem. 2008;283:18177–86.
Sragovich S, Amram N, Yeheskel A, Gozes I. VIP/PACAP-based drug development: the ADNP/NAP-derived mirror peptides SKIP and D-SKIP exhibit distinctive in vivo and in silico effects. Front Cell Neurosci. 2019;13:589.
Sragovich S, Malishkevich A, Piontkewitz Y, Giladi E, Touloumi O, Lagoudaki R, et al. The autism/neuroprotection-linked ADNP/NAP regulate the excitatory glutamatergic synapse. Transl Psychiatry. 2019;9:2.
Jaworski J, Hoogenraad CC, Akhmanova A. Microtubule plus-end tracking proteins in differentiated mammalian cells. Int J Biochem cell Biol. 2008;40:619–37.
Mukaetova-Ladinska EB, Arnold H, Jaros E, Perry R, Perry E. Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol Appl Neurobiol. 2004;30:615–23.
Mandel S, Spivak-Pohis I, Gozes I. ADNP differential nucleus/cytoplasm localization in neurons suggests multiple roles in neuronal differentiation and maintenance. J Mol Neurosci. 2008;35:127–41.
Garbern JY, Neumann M, Trojanowski JQ, Lee VM, Feldman G, Norris JW, et al. A mutation affecting the sodium/proton exchanger, SLC9A6, causes mental retardation with tau deposition. Brain. 2010;133:1391–402.
Schirer Y, Malishkevich A, Ophir Y, Lewis J, Giladi E, Gozes I. Novel marker for the onset of frontotemporal dementia: early increase in activity-dependent neuroprotective protein (ADNP) in the face of Tau mutation. PloS ONE. 2014;9:e87383.
Mahmood SR, Xie X, Hosny El Said N, Venit T, Gunsalus KC, Percipalle P. beta-actin dependent chromatin remodeling mediates compartment level changes in 3D genome architecture. Nat Commun. 2021;12:5240.
Latapy C, Rioux V, Guitton MJ, Beaulieu JM. Selective deletion of forebrain glycogen synthase kinase 3beta reveals a central role in serotonin-sensitive anxiety and social behaviour. Philos Trans R Soc Lond Ser B Biol Sci. 2012;367:2460–74.
Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91.
Teyra J, Huang H, Jain S, Guan X, Dong A, Liu Y, et al. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure. 2017;25:1598–610.e3.
Kurochkina N, Guha U. SH3 domains: modules of protein-protein interactions. Biophys Rev. 2013;5:29–39.
Kohlenberg TM, Trelles MP, McLarney B, Betancur C, Thurm A, Kolevzon A. Psychiatric illness and regression in individuals with Phelan-McDermid syndrome. J Neurodev Disord. 2020;12:7.
Alcalay RN, Giladi E, Pick CG, Gozes I. Intranasal administration of NAP, a neuroprotective peptide, decreases anxiety-like behavior in aging mice in the elevated plus maze. Neurosci Lett. 2004;361:128–31.
Zisapel N, Levi M, Gozes I. Tubulin: an integral protein of mammalian synaptic vesicle membranes. J Neurochem. 1980;34:26–32.
Hofstein R, Hershkowitz M, Gozes I, Samuel D. The characterization and phosphorylation of an actin-like protein in synaptosomal membranes. Biochim Biophys Acta. 1980;624:153–62.
MacGillavry HD, Kerr JM, Kassner J, Frost NA, Blanpied TA. Shank-cortactin interactions control actin dynamics to maintain flexibility of neuronal spines and synapses. Eur J Neurosci. 2016;43:179–93.
Amal H, Barak B, Bhat V, Gong G, Joughin BA, Wang X, et al. Shank3 mutation in a mouse model of autism leads to changes in the S-nitroso-proteome and affects key proteins involved in vesicle release and synaptic function. Mol Psychiatry. 2020;25:1835–48.
Cosgrave AS, McKay JS, Morris R, Quinn JP, Thippeswamy T. Nitric oxide regulates activity-dependent neuroprotective protein (ADNP) in the dentate gyrus of the rodent model of kainic acid-induced seizure. J Mol Neurosci. 2009;39:9–21.
Ashur-Fabian O, Giladi E, Furman S, Steingart RA, Wollman Y, Fridkin M, et al. Vasoactive intestinal peptide and related molecules induce nitrite accumulation in the extracellular milieu of rat cerebral cortical cultures. Neurosci Lett. 2001;307:167–70.
Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Mol Autism. 2014;5:30.
Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–8.
Acknowledgements
We are grateful to Tal Barak and Elad Malikov for their diligent work with the mutant Shank3 model mice and Prof. R. Frank Kooy for the ADNP patient-derived lymphoblastoid cell lines. This work was partially supported by grants to Prof. IG from the European Research Area Network (ERA-NET) Neuron ADNPinMED, Drs. Ronith and Armand Stemmer and Arthur Gerbi (French Friends of Tel Aviv University), Holly and Jonathan Strelzik (American Friends of Tel Aviv University) and Anne and Alex Cohen (Canadian Friends of Tel Aviv University). IG and BB are further supported by Sagol School of Neuroscience Interdisciplinary Partnership Grant (SNIP). IG is Director of the Elton Laboratory for Molecular Neuroendocrinology and the former first incumbent of the Lily and Avraham Gildor Chair for the Investigation of Growth Factors. This work is in partial fulfillment of Ph.D. thesis requirements at the Miriam and Sheldon G. Adelson Graduate School, Sackler Faculty of Medicine, Tel Aviv University (MG), who was also generously supported by a Neubauer Family Foundation Student Scholarship. This work is also in partial fulfillment of M.Sc. thesis requirements at the Miriam and Sheldon G. Adelson Graduate School (AL) and at the Sagol School of Neuroscience, Tel Aviv University (IB-H-H).
Author information
Authors and Affiliations
Contributions
Together with IG, YI-P planned and performed all the cell culture experiments (Figs. 1–4), and wrote an initial, in depth, draft. MG performed biochemical experiments outlined in Fig. 5. IB-H-H performed animal behavior experiments. AL performed biochemical experiments outlined in Fig. 5. NB helped with the biochemical experiments. IF, GL, SSr, GK, and EG helped with all animal experiments. SSh performed in silico analyses (Fig. 5). Homozygous Shank3 ASD-linked InsG3680 mutated mice were housed and tested under the supervision of BB. IG orchestrated and supervised the entire project and wrote the paper with data analysis and editorial remarks from all contributing authors.
Corresponding author
Ethics declarations
Competing interests
NAP (CP201, davunetide) use is under patent protection (US patent nos. US7960334, US8618043, and USWO2017130190A1) (IG), PCT/IL2020/051010 (IG) and provisional patent applications (IG inventor and contributing scientists, YI-P, MG, IB-H-H, SS, EG). Davunetide is exclusively licensed to ATED Therapeutics LTD (IG, Co-Founder and Chief Scientific Officer).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ivashko-Pachima, Y., Ganaiem, M., Ben-Horin-Hazak, I. et al. SH3- and actin-binding domains connect ADNP and SHANK3, revealing a fundamental shared mechanism underlying autism. Mol Psychiatry 27, 3316–3327 (2022). https://doi.org/10.1038/s41380-022-01603-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01603-w
This article is cited by
-
Clinical impact and in vitro characterization of ADNP variants in pediatric patients
Molecular Autism (2024)
-
ADNP dysregulates methylation and mitochondrial gene expression in the cerebellum of a Helsmoortel–Van der Aa syndrome autopsy case
Acta Neuropathologica Communications (2024)
-
Longitudinal Genotype-Phenotype (Vineland Questionnaire) Characterization of 15 ADNP Syndrome Cases Highlights Mutated Protein Length and Structural Characteristics Correlation with Communicative Abilities Accentuated in Males
Journal of Molecular Neuroscience (2024)
-
Chromatin remodeler Activity-Dependent Neuroprotective Protein (ADNP) contributes to syndromic autism
Clinical Epigenetics (2023)
-
A novel genomic mutation in ADNP leading to intellectual disability
European Journal of Human Genetics (2023)