Abstract
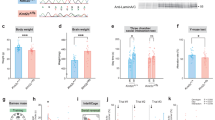
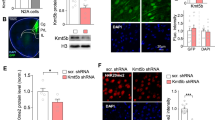
Large-scale genetic studies have revealed that the most prominent genes disrupted in autism are chromatin regulators mediating histone methylation/demethylation, suggesting the central role of epigenetic dysfunction in this disorder. Here, we show that histone lysine 4 dimethylation (H3K4me2), a histone mark linked to gene activation, is significantly decreased in the prefrontal cortex (PFC) of autistic human patients and mutant mice with the deficiency of top-ranking autism risk factor Shank3 or Cul3. A brief treatment of the autism models with highly potent and selective inhibitors of the H3K4me2 demethylase LSD1 (KDM1A) leads to the robust rescue of core symptoms of autism, including social deficits and repetitive behaviors. Concomitantly, LSD1 inhibition restores NMDA receptor function in PFC and AMPA receptor-mediated currents in striatum of Shank3-deficient mice. Genome-wide RNAseq and ChIPseq reveal that treatment of Shank3-deficient mice with the LSD1 inhibitor restores the expression and H3K4me2 occupancy of downregulated genes enriched in synaptic signaling and developmental processes. The immediate early gene tightly linked to neuronal plasticity, Egr1, is on the top list of rescued genes. The diminished transcription of Egr1 is recapitulated in PFC of autistic human patients. Overexpression of Egr1 in PFC of Shank3-deficient mice ameliorates social preference deficits. These results have for the first time revealed an important role of H3K4me2 abnormality in ASD pathophysiology, and the therapeutic potential of targeting H3K4me2 demethylase LSD1 or the downstream molecule Egr1 for ASD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNAseq and ChIPseq data generated in this study have been deposited in the GEO public repository under accession code GSE193380.
References
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
Coe BP, Stessman HAF, Sulovari A, Geisheker MR, Bakken TE, Lake AM, et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51:106–16.
Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–26.
Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–20.
Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol. 2019;20:625–41.
Tunovic S, Barkovich J, Sherr EH, Slavotinek AM. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am J Med Genet A. 2014;164A:1744–9.
Shen E, Shulha H, Weng Z, Akbarian S. Regulation of histone H3K4 methylation in brain development and disease. Philos Trans R Soc Biol Sci 2014;369:20130514.
Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69:314–24.
Betancur C, Buxbaum JD. SHANK3 haploinsufficiency: a “common” but underdiagnosed highly penetrant monogenic cause of autism spectrum disorders. Mol Autism. 2013;4:17.
Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10:e1004580.
Qin L, Ma K, Wang ZJ, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–75.
Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like deficits in Shank3-Deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–13.
Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42.
Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Escamilla CO, Weaver TP, et al. Novel Shank3 mutant exhibits behaviors with face validity for autism and altered striatal and hippocampal function. Autism Res. 2017;10:42–65.
Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat Commun. 2016;7:11459.
Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–108.
Wang ZJ, Zhong P, Ma K, Seo JS, Yang F, Hu Z, et al. Amelioration of autism-like social deficits by targeting histone methyltransferases EHMT1/2 in Shank3-deficient mice. Mol Psychiatry. 2020;25:2517–33.
Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–402.
Nair SS, Li DQ, Kumar R. A core chromatin remodeling factor instructs global chromatin signaling through multivalent reading of nucleosome codes. Mol Cell. 2013;49:704–18.
Meier K, Brehm A. Chromatin regulation: how complex does it get? Epigenetics. 2014;9:1485–95.
Yan Z, Rein B. Mechanisms of synaptic transmission dysregulation in the prefrontal cortex: pathophysiological implications. Mol Psychiatry. 2021. https://doi.org/10.1038/s41380-021-01092-3. Online ahead of print.
Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. N. Engl J Med. 2014;370:1209–19.
Cusan M, Cai SF, Mohammad HP, Krivtsov A, Chramiec A, Loizou E, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood. 2018;131:1730–42.
Rein B, Ma K, Yan Z. A standardized social preference protocol for measuring social deficits in mouse models of autism. Nat Protoc. 2020;15:3464–77.
Maes T, Mascaro C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, et al. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell. 2018;33:495–511.
Rapanelli M, Tan T, Wang W, Wang X, Wang ZJ, Zhong P, et al. Behavioral, circuitry, and molecular aberrations by region-specific deficiency of the high-risk autism gene Cul3. Mol Psychiatry. 2021;26:1491–504.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50.
Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–55.
Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–20.
Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–5.
Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, Gupta N, et al. Autism-associated insertion mutation (InsG) of Shank3 exon 21 causes impaired synaptic transmission and behavioral deficits. J Neurosci. 2015;35:9648–65.
Wang W, Li C, Chen Q, van der Goes MS, Hawrot J, Yao AY, et al. Striatopallidal dysfunction underlies repetitive behavior in Shank3-deficient model of autism. J Clin Invest. 2017;127:1978–90.
Duclot F, Kabbaj M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front Behav Neurosci. 2017;11:35.
Mukai J, Cannavo E, Crabtree GW, Sun Z, Diamantopoulou A, Thakur P, et al. Recapitulation and reversal of schizophrenia-related phenotypes in Setd1a-deficient mice. Neuron. 2019;104:471–87.
Kundakovic M, Jiang Y, Kavanagh DH, Dincer A, Brown L, Pothula V, et al. Practical guidelines for high-resolution epigenomic profiling of nucleosomal histones in postmortem human brain tissue. Biol Psychiatry. 2017;81:162–70.
Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364:685–9.
Maze I, Covington HE 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6.
Hamilton PJ, Burek DJ, Lombroso SI, Neve RL, Robison AJ, Nestler EJ, et al. Cell-type-specific epigenetic editing at the fosb gene controls susceptibility to social defeat stress. Neuropsychopharmacology. 2018;43:272–84.
Ma K, Qin L, Matas E, Duffney LJ, Liu A, Yan Z. Histone deacetylase inhibitor MS-275 restores social and synaptic function in a Shank3-deficient mouse model of autism. Neuropsychopharmacology. 2018;43:1779–88.
Qin L, Ma K, Yan Z. Chemogenetic activation of prefrontal cortex in Shank3-deficient mice ameliorates social deficits, NMDAR hypofunction, and Sgk2 downregulation. iScience. 2019;17:24–35.
Qin L, Ma K, Yan Z. Rescue of histone hypoacetylation and social deficits by ketogenic diet in a Shank3 mouse model of autism. Neuropsychopharmacology. 2021. https://doi.org/10.1038/s41386-021-01212-1. Online ahead of print.
Graybiel AM. Habits, rituals, and the evaluative brain. Annu Rev Neurosci. 2008;31:359–87.
Ahmari SE, Spellman T, Douglass NL, Kheirbek MA, Simpson HB, Deisseroth K, et al. Repeated cortico-striatal stimulation generates persistent OCD-like behavior. Science. 2013;340:1234–9.
Hadjas LC, Schartner MM, Cand J, Creed MC, Pascoli V, Lüscher C, et al. Projection-specific deficits in synaptic transmission in adult Sapap3-knockout mice. Neuropsychopharmacology. 2020;45:2020–9.
Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211.
Koldamova R, Schug J, Lefterova M, Cronican AA, Fitz NF, Davenport FA, et al. Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration. Neurobiol Dis. 2014;63:107–14.
Covington HE 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30:16082–90.
Kimoto S, Bazmi HH, Lewis DA. Lower expression of glutamic acid decarboxylase 67 in the prefrontal cortex in schizophrenia: contribution of altered regulation by Zif268. Am J Psychiatry. 2014;171:969–78.
Yamada K, Gerber DJ, Iwayama Y, Ohnishi T, Ohba H, Toyota T, et al. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc Natl Acad Sci. 2007;104:2815–20.
Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, et al. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci. 2016;113:3651–6.
Dossat AM, Jourdi H, Wright KN, Strong CE, Sarkar A, Kabbaj M. Viral-mediated Zif268 expression in the prefrontal cortex protects against gonadectomy-induced working memory, long-term memory, and social interaction deficits in male rats. Neuroscience. 2017;340:243–57.
Stack A, Carrier N, Dietz D, Hollis F, Sorenson J, Kabbaj M. Sex differences in social interaction in rats: role of the immediate-early gene zif268. Neuropsychopharmacology. 2010;35:570–80.
Yang Y, Shu X, Liu D, Shang Y, Wu Y, Pei L, et al. EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron. 2012;73:774–88.
Acknowledgements
We thank Dr. Craig Powell at University of Alabama at Birmingham for kindly providing breeding pairs of Shank3E13 mice. We thank Dr. Mohamed Kabbaj at Florida State University for kindly providing Egr1 AAV and GFP AAV. We also thank Xiaoqing Chen for excellent technical support and Luciana R. Frick for taking some confocal images. This work was supported by: Nancy Lurie Marks Family Foundation (ZY), National Institutes of Health (MH112237, MH126443) (ZY), National Center for Advancing Translational Sciences of the National Institutes of Health (KL2TR001413) (MR), and National Institutes of Health (MH104341, MH117790) (SA).
Author information
Authors and Affiliations
Contributions
MR performed some behavioral, immunocytochemical, biochemical experiments, bioinformatic analyses, and wrote the draft. JW and FY performed bioinformatic analyses. KM performed behavioral experiments. PZ carried out electrophysiological experiments. RP and MK performed some behavioral and biochemical assays. LQ, BR, Z-JW carried out qPCR or immunocytochemical experiments. BK, BJ, and LC extracted neuronal nuclei for ChIPseq experiments. SA oversaw ChIPseq studies. ZY designed experiments, supervised the project and wrote the paper. None of the authors have the financial interest related to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Rapanelli, M., Williams, J.B., Ma, K. et al. Targeting histone demethylase LSD1 for treatment of deficits in autism mouse models. Mol Psychiatry 27, 3355–3366 (2022). https://doi.org/10.1038/s41380-022-01508-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01508-8