Abstract
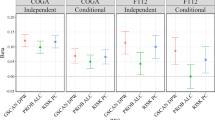
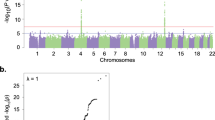
Runs of homozygosity (ROH) arise when an individual inherits two copies of the same haplotype segment. While ROH are ubiquitous across human populations, Native populations—with shared parental ancestry arising from isolation and endogamy—can carry a substantial enrichment for ROH. We have been investigating genetic and environmental risk factors for alcohol use disorders (AUD) in a group of American Indians (AI) who have higher rates of AUD than the general U. S. population. Here we explore whether ROH might be associated with incidence and severity of AUD in this admixed AI population (n = 742) that live on geographically contiguous reservations, using low-coverage whole genome sequences. We have found that the genomic regions in the ROH that were identified in this population had significantly elevated American Indian heritage compared with the rest of the genome. Increased ROH abundance and ROH burden are likely risk factors for AUD severity in this AI population, especially in those diagnosed with severe and moderate AUD. The association between ROH and AUD was mostly driven by ROH of moderate lengths between 1 and 2 Mb. An ROH island on chromosome 1p32.3 and a rare ROH pool on chromosome 3p12.3 were found to be significantly associated with AUD severity. They contain genes involved in lipid metabolism, oxidative stress and inflammatory responses; and OSBPL9 was found to reside on the consensus part of the ROH island. These data demonstrate that ROH are associated with risk for AUD severity in this AI population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data and code availability
In accordance with the wishes of the tribes no sharing of the AI data are possible. All analysis codes were written in R and are available upon requests.
References
Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. Epidemiology of DSM-5 alcohol use disorder: Results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72:757–66.
Compton WM, Thomas YF, Stinson FS, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV drug abuse and dependence in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2007;64:566–76.
Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry. 2013;170:154–64.
Edenberg HJ, McClintick JN. Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: a critical review. Alcohol Clin Exp Res. 2018;42:2281–97.
Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10:487–94.
Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–9.
Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, et al. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry. 2017;22:1359.
Hart AB, Kranzler HR. Alcohol dependence genetics: lessons learned from genome-wide association studies (GWAS) and post-GWAS analyses. Alcohol: Clin Exp Res. 2015;39:1312–27.
Peng Q, Gizer IR, Libiger O, Bizon C, Wilhelmsen KC, Schork NJ, et al. Association and ancestry analysis of sequence variants in ADH and ALDH using alcohol-related phenotypes in a Native American community sample. Am J Med Genet Part B: Neuropsychiatr Genet. 2014;165:673–83.
Peng Q, Gizer IR, Wilhelmsen KC, Ehlers CL. Associations between genomic variants in alcohol dehydrogenase genes and alcohol symptomatology in American Indians and European Americans: distinctions and convergence. Alcohol: Clin Exp Res. 2017;41:1695–704.
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, et al. Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2018;176:107–18.
Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499.
Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 2017;22:1376.
Peng Q, Bizon C, Gizer IR, Wilhelmsen KC, Ehlers CL. Genetic loci for alcohol-related life events and substance-induced affective symptoms: indexing the “dark side” of addiction. Transl Psychiatry. 2019;9:71.
Minster RL, Hawley NL, Su C-T, Sun G, Kershaw EE, Cheng H, et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat Genet. 2016;48:1049–54.
Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–5.
Ehlers CL, Wilhelmsen KC. Genomic screen for substance dependence and body mass index in southwest California Indians. Genes Brain Behav. 2007;6:184–91.
Mooney JA, Huber CD, Service S, Sul JH, Marsden CD, Zhang Z, et al. Understanding the hidden complexity of Latin American population isolates. Am J Hum Genet. 2018;103:707–26.
Leske S, Harris MG, Charlson FJ, Ferrari AJ, Baxter AJ, Logan JM, et al. Systematic review of interventions for Indigenous adults with mental and substance use disorders in Australia, Canada, New Zealand and the United States. Aust NZ J Psychiatry. 2016;50:1040–54.
Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19:220.
Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–95.
Szpiech Zachary A, Xu J, Pemberton Trevor J, Peng W, Zöllner S, Rosenberg Noah A, et al. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet. 2013;93:90–102.
Szpiech ZA, Mak ACY, White MJ, Hu D, Eng C, Burchard EG, et al. Ancestry-dependent enrichment of deleterious homozygotes in runs of homozygosity. Am J Hum Genet. 2019;105:747–62.
Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, et al. A simple genetic architecture underlies morphological variation in dogs. PLOS Biol. 2010;8:e1000451.
Kim E-S, Cole JB, Huson H, Wiggans GR, Van Tassell CP, Crooker BA, et al. Effect of artificial selection on runs of homozygosity in U.S. Holstein Cattle. PLOS One. 2013;8:e80813.
Qanbari S, Pimentel ECG, Tetens J, Thaller G, Lichtner P, Sharifi AR, et al. A genome-wide scan for signatures of recent selection in Holstein cattle. Anim Genet. 2010;41:377–89.
Ablondi M, Viklund Å, Lindgren G, Eriksson S, Mikko S. Signatures of selection in the genome of Swedish warmblood horses selected for sport performance. BMC Genomics. 2019;20:717.
Metzger J, Karwath M, Tonda R, Beltran S, Águeda L, Gut M, et al. Runs of homozygosity reveal signatures of positive selection for reproduction traits in breed and non-breed horses. BMC Genomics. 2015;16:764.
Purfield DC, McParland S, Wall E, Berry DP. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PloS One. 2017;12:e0176780-e.
Peripolli E, Munari DP, Silva MVGB, Lima ALF, Irgang R, Baldi F. Runs of homozygosity: current knowledge and applications in livestock. Anim Genet. 2017;48:255–71.
McQuillan R, Eklund N, Pirastu N, Kuningas M, McEvoy BP, Esko T, et al. Evidence of inbreeding depression on human height. PLOS Genet. 2012;8:e1002655.
Hu H, Kahrizi K, Musante L, Fattahi Z, Herwig R, Hosseini M, et al. Genetics of intellectual disability in consanguineous families. Mol Psychiatry. 2018;24:1027–39.
Gandin I, Faletra F, Faletra F, Carella M, Pecile V, Ferrero GB, et al. Excess of runs of homozygosity is associated with severe cognitive impairment in intellectual disability. Genet Med. 2014;17:396.
Howrigan DP, Simonson MA, Davies G, Harris SE, Tenesa A, Starr JM, et al. Genome-wide autozygosity is associated with lower general cognitive ability. Mol Psychiatry. 2015;21:837.
Howrigan DP, Simonson MA, Davies G, Harris SE, Tenesa A, Starr JM, et al. Genome-wide autozygosity is associated with lower general cognitive ability. Mol Psychiatry. 2016;21:837–43.
Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci. 2007;104:19942.
Keller MC, Simonson MA, Ripke S, Neale BM, Gejman PV, Howrigan DP, et al. Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor. PLOS Genet. 2012;8:e1002656.
Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–79.
Ghani M, Reitz C, Cheng R, et al. Association of long runs of homozygosity with alzheimer disease among african american individuals. JAMA Neurol. 2015;72:1313–23.
Nalls MA, Guerreiro RJ, Simon-Sanchez J, Bras JT, Traynor BJ, Gibbs JR, et al. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer’s disease. Neurogenetics. 2009;10:183–90.
Clark DW, Okada Y, Moore KHS, Mason D, Pirastu N, Gandin I, et al. Associations of autozygosity with a broad range of human phenotypes. Nat Commun. 2019;10:4957.
Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. Am J Psychiatry. 2004;161:1204–10.
Abdellaoui A, Hottenga J-J, Willemsen G, Bartels M, van Beijsterveldt T, Ehli EA, et al. Educational attainment influences levels of homozygosity through migration and assortative mating. PLOS ONE. 2015;10:e0118935.
Johnson EC, Evans LM, Keller MC. Relationships between estimated autozygosity and complex traits in the UK Biobank. PLOS Genet. 2018;14:e1007556.
Ehlers CL, Gizer IR, Gilder DA, Wilhelmsen KC. Linkage analyses of stimulant dependence, craving, and heavy use in American Indians. Am J Med Genet Part B. 2011;156B:772–80.
Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG. Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in southwest California Indians. Alcohol Clin Exp Res. 2004;28:1481–6.
Kalton G, Anderson DW. Sampling rare populations. J R Stat Soc. 1986;149:65–82.
Muhib FB, Lin LS, Stueve A, Miller RL, Ford WL, Johnson WD, et al. A venue-based method for sampling hard-to-reach populations. Public Health Rep. 2001;116:216–22.
Heckathorn DD. Respondent-driven sampling: a new approach to the study of hidden populations*. Soc Probl. 2014;44:174–99.
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr., et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–58.
Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA–a comparison with the SCAN. Addiction. 1999;94:1361–70.
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013.
Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry. 2003;160:41–6.
Hesselbrock VM, Segal B, Hesselbrock MN. Alcohol dependence among Alaska Natives entering alcoholism treatment: a gender comparison. J Stud Alcohol. 2000;61:150–6.
Gilder DA, Wall TL, Ehlers CL. Comorbidity of select anxiety and affective disorders with alcohol dependence in southwest California Indians. Alcohol Clin Exp Res. 2004;28:1805–13.
Schuckit MA, Smith TL, Anthenelli R, Irwin M. Clinical course of alcoholism in 636 male inpatients. Am J Psychiatry. 1993;150:786–92.
Ehlers CL, Stouffer GM, Corey L, Gilder DA. The clinical course of DSM-5 alcohol use disorders in young adult native and Mexican Americans. Am J Addictions. 2015;24:713–21.
Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, et al. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15:85.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.0.1–33.
Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–51.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Ceballos FC, Hazelhurst S, Ramsay M. Assessing runs of Homozygosity: a comparison of SNP Array and whole genome sequence low coverage data. BMC Genomics. 2018;19:106.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Chen H, Wang C, Conomos Matthew P, Stilp Adrienne M, Li Z, Sofer T, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–66.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statis Soc B. 1995;57:289–300.
Leisch F. FlexMix: a general framework for finite mixture models and latent class regression in R. J Stat Softw. 2004;1:2004.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018;47:D886–94.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen Egon L, Bohler A, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–94.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2013;42:D1001–6.
Nothnagel M, Lu TT, Kayser M, Krawczak M. Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans. Hum Mol Genet. 2010;19:2927–35.
GTEx Consortium T, Aguet F, Brown AA, Castel SE, Davis JR, He Y, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21.
Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–57.
Velez Edwards DR, Naj AC, Monda K, North KE, Neuhouser M, Magvanjav O, et al. Gene-environment interactions and obesity traits among postmenopausal African-American and Hispanic women in the Women’s Health Initiative SHARe Study. Hum Genet. 2013;132:323–36.
Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–69.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Matoba N, Akiyama M, Ishigaki K, Kanai M, Takahashi A, Momozawa Y, et al. GWAS of 165,084 Japanese individuals identified nine loci associated with dietary habits. Nat Hum Behav. 2020;4:308–16.
Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31–41.
Sun Y, Chang S, Liu Z, Zhang L, Wang F, Yue W, et al. Identification of novel risk loci with shared effects on alcoholism, heroin, and methamphetamine dependence. Molecular Psychiatry. 2019. https://doi.org/10.1038/s41380-019-0497-y.
Whitfield JB, Zhu G, Madden PAF, Montgomery GW, Heath AC, Martin NG. Biomarker and genomic risk factors for liver function test abnormality in hazardous drinkers. Alcohol, Clin Exp Res. 2019;43:473–82.
Mutemberezi V, Buisseret B, Masquelier J, Guillemot-Legris O, Alhouayek M, Muccioli GG. Oxysterol levels and metabolism in the course of neuroinflammation: insights from in vitro and in vivo models. J Neuroinflammation. 2018;15:74.
Sawyer KS, Adra N, Salz DM, Kemppainen MI, Ruiz SM, Harris GJ, et al. Hippocampal subfield volumes in abstinent men and women with a history of alcohol use disorder. PLOS ONE. 2020;15:e0236641.
Maller JJ, Broadhouse K, Rush AJ, Gordon E, Koslow S, Grieve SM. Increased hippocampal tail volume predicts depression status and remission to anti-depressant medications in major depression. Mol Psychiatry. 2018;23:1737–44.
Skillbäck T, Farahmand BY, Rosén C, Mattsson N, Nägga K, Kilander L, et al. Cerebrospinal fluid tau and amyloid-β1-42 in patients with dementia. Brain. 2015;138:2716–31.
Kalinin S, González-Prieto M, Scheiblich H, Lisi L, Kusumo H, Heneka MT, et al. Transcriptome analysis of alcohol-treated microglia reveals downregulation of beta amyloid phagocytosis. J Neuroinflammation. 2018;15:141.
Olex AL, Hiltbold EM, Leng X, Fetrow JS. Dynamics of dendritic cell maturation are identified through a novel filtering strategy applied to biological time-course microarray replicates. BMC Immunol. 2010;11:41.
Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257.
Lawrimore CJ, Crews FT. Ethanol, TLR3, and TLR4 agonists have unique innate immune responses in neuron-like SH-SY5Y and microglia-Like BV2. Alcohol: Clin Exp Res. 2017;41:939–54.
Warden AS, Azzam M, DaCosta A, Mason S, Blednov YA, Messing RO, et al. Toll-like receptor 3 activation increases voluntary alcohol intake in C57BL/6J male mice. Brain Behav Immun. 2019;77:55–65.
Karlsson G, Liu Y, Larsson J, Goumans M-J, Lee J-S, Thorgeirsson SS, et al. Gene expression profiling demonstrates that TGF-β1 signals exclusively through receptor complexes involving Alk5 and identifies targets of TGF-β signaling. Physiol Genom. 2005;21:396–403.
Breitkopf K, Haas S, Wiercinska E, Singer MV, Dooley S. Anti-TGF-β Strategies for the treatment of chronic liver disease. Alcohol: Clin Exp Res. 2005;29:121S–31S.
Kim Y-K, Lee BC, Ham BJ, Yang B-H, Roh S, Choi J, et al. Increased transforming growth factor-beta1 in alcohol dependence. J Korean Med Sci. 2009;24:941–4.
Siegenthaler JA, Miller MW. Transforming growth factor β1 modulates cell migration in rat cortex: effects of ethanol. Cereb Cortex. 2004;14:791–802.
Steiner JL, Lang CH. Alcohol, adipose tissue and lipid dysregulation. Biomolecules. 2017;7:16.
Jones SE, Lane JM, Wood AR, van Hees VT, Tyrrell J, Beaumont RN, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10:343.
Hoffmann TJ, Choquet H, Yin J, Banda Y, Kvale MN, Glymour M, et al. A large multiethnic genome-wide association study of adult body mass index identifies novel loci. Genetics. 2018;210:499.
Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Acknowledgements
We would like to acknowledge and thank all of our American Indian participants, and the following people for their roles in (1) the genotyping effort: Kirk Wilhelmsen, Scott Chasse, Piotr Mieczkowski, Ewa Patrycja Malc, Joshua Sailsbery, Phil Owens, and Chris Bizon; and (2) recruiting participants, and collection and preparation of the clinical data: David Gilder, Corinne Kim, Evie Phillips, Phillip Lau, and Derek Wills. This work was supported by the National Institutes of Health (NIH): National Institute on Alcohol Abuse and Alcoholism (NIAAA) K25 AA025095 to QP; NIAAA R01 AA027316 to CLE; National Institute on Drug Abuse (NIDA) R01 DA030976 to CLE. NIAAA and NIDA had no further role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Peng, Q., Ehlers, C.L. Long tracks of homozygosity predict the severity of alcohol use disorders in an American Indian population. Mol Psychiatry 26, 2200–2211 (2021). https://doi.org/10.1038/s41380-020-00989-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00989-9
This article is cited by
-
Genetic factors associated with suicidal behaviors and alcohol use disorders in an American Indian population
Molecular Psychiatry (2024)