Abstract
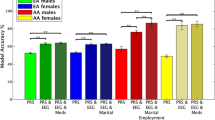
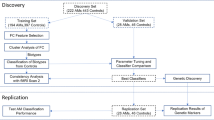
Predictive models have succeeded in distinguishing between individuals with Alcohol use Disorder (AUD) and controls. However, predictive models identifying who is prone to develop AUD and the biomarkers indicating a predisposition to AUD are still unclear. Our sample (n = 656) included offspring and non-offspring of European American (EA) and African American (AA) ancestry from the Collaborative Study of the Genetics of Alcoholism (COGA) who were recruited as early as age 12 and were unaffected at first assessment and reassessed years later as AUD (DSM-5) (n = 328) or unaffected (n = 328). Machine learning analysis was performed for 220 EEG measures, 149 alcohol-related single nucleotide polymorphisms (SNPs) from a recent large Genome-wide Association Study (GWAS) of alcohol use/misuse and two family history (mother DSM-5 AUD and father DSM-5 AUD) features using supervised, Linear Support Vector Machine (SVM) classifier to test which features assessed before developing AUD predict those who go on to develop AUD. Age, gender, and ancestry stratified analyses were performed. Results indicate significant and higher accuracy rates for the AA compared with the EA prediction models and a higher model accuracy trend among females compared with males for both ancestries. Combined EEG and SNP features model outperformed models based on only EEG features or only SNP features for both EA and AA samples. This multidimensional superiority was confirmed in a follow-up analysis in the AA age groups (12–15, 16–19, 20–30) and EA age group (16–19). In both ancestry samples, the youngest age group achieved higher accuracy score than the two other older age groups. Maternal AUD increased the model’s accuracy in both ancestries’ samples. Several discriminative EEG measures and SNPs features were identified, including lower posterior gamma, higher slow wave connectivity (delta, theta, alpha), higher frontal gamma ratio, higher beta correlation in the parietal area, and 5 SNPs: rs4780836, rs2605140, rs11690265, rs692854, and rs13380649. Results highlight the significance of sampling uniformity followed by stratified (e.g., ancestry, gender, developmental period) analysis, and wider selection of features, to generate better prediction scores allowing a more accurate estimation of AUD development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Patrick ME, Schulenberg JE. Prevalence and predictors of adolescent alcohol use and binge drinking in the United States. Alcohol Res. 2013;35:193–200.
Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40.
Whelan R, Watts R, Orr CA, Althoff RR, Artiges E, Banaschewski T, et al. Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature. 2014;512:185–9.
Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. A hybrid machine learning method for fusing fmri and genetic data: combining both improves classification of schizophrenia. Front Hum Neurosci. 2010;4:192.
Librenza-Garcia D, Kotzian BJ, Yang J, Mwangi B, Cao B, Pereira Lima LN, et al. The impact of machine learning techniques in the study of bipolar disorder: a systematic review. Neurosci Biobehav Rev. 2017;80:538–54.
Sacchet MD, Prasad G, Foland-Ross LC, Thompson PM, Gotlib IH. Support vector machine classification of major depressive disorder using diffusion-weighted neuroimaging and graph theory. Front Psychiatry. 2015;6:21.
Bi J, Sun J, Wu Y, Tennen H, Armeli S. A machine learning approach to college drinking prediction and risk factor identification. ACM Trans Intell Syst Technol. 2013;4:1–24.
Shim M, Hwang HJ, Kim DW, Lee SH, Im CH. Machine-learning-based diagnosis of schizophrenia using combined sensor-level and source-level EEG features. Schizophr Res. 2016;176:314–9.
Acharya UR, Sree SV, Chattopadhyay S, Suri JS. Automated diagnosis of normal and alcoholic EEG signals. Int J Neural Syst. 2012;22:1250011.
Mumtaz W, Vuong PL, Xia LK, Malik AS, Bin ABD, Rashidb R. Automatic diagnosis of alcohol use disorder using EEG features. Knowl-Based Syst. 2016;105:48–59.
Mumtaz W, Vuong P, Xia LK, Malik A, Bin Abd Rashid R. An EEG-based machine learning method to screen alcohol use disorder. Cogn Neurodyn. 2017;11:161–71.
Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 2017;22:1376–84.
Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, et al. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry. 2017;22:1359–67.
Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19:41–9.
Polimanti R, Zhang H, Smith AH, Zhao H, Farrer LA, Kranzler HR, et al. Genome-wide association study of body mass index in subjects with alcohol dependence. Addict Biol. 2017;22:535–49.
Meyers JL, Zhang J, Wang JC, Su J, Kuo SI, Kapoor M, et al. An endophenotype approach to the genetics of alcohol dependence: a genome wide association study of fast beta EEG in families of African ancestry. Mol Psychiatry. 2017;22:1767–75.
Pierce TW, Watson TD, King JS, Kelly SP, Pribram KH. Age differences in factor analysis of EEG. Brain Topogr. 2003;16:19–27.
Zappasodi F, Marzetti L, Olejarczyk E, Tecchio F, Pizzella V. Age-related changes in electroencephalographic signal complexity. PLoS ONE. 2015;10:e0141995.
Chorlian DB, Rangaswamy M, Manz N, Kamarajan C, Pandey AK, Edenberg H, et al. Gender modulates the development of theta event related oscillations in adolescents and young adults. Behav Brain Res. 2015;292:342–52.
Ngun TC, Ghahramani N, Sanchez FJ, Bocklandt S, Vilain E. The genetics of sex differences in brain and behavior. Front Neuroendocrinol. 2011;32:227–46.
Ali-Khan SE, Krakowski T, Tahir R, Daar AS. The use of race, ethnicity and ancestry in human genetic research. Hugo J. 2011;5:47–63.
Sankar P, Cho MK. Genetics: toward a new vocabulary of human genetic variation. Science. 2002;298:1337–8.
Li L, Rakitsch B, Borgwardt K. ccSVM: correcting Support Vector Machines for confounding factors in biological data classification. Bioinformatics. 2011;27:i342–348.
Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, et al. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108:244–50.
Edenberg HJ, Bierut LJ, Boyce P, Cao M, Cawley S, Chiles R, et al. Description of the data from the Collaborative Study on the Genetics of Alcoholism (COGA) and single-nucleotide polymorphism genotyping for Genetic Analysis Workshop 14. BMC Genet. 2005;6(Suppl 1):S2.
Reich T. A genomic survey of alcohol dependence and related phenotypes: results from the Collaborative Study on the Genetics of Alcoholism (COGA). Alcohol Clin Exp Res. 1996;20(8 Suppl):133A–137A.
Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2:861–3.
Macleod S, Appleton RE. Neurological disorders presenting mainly in adolescence. Arch Dis Child. 2007;92:170–5.
Klem GH, Luders HO, Jasper HH, Elger C. The ten-twenty electrode system of the International Federation. The International Federation of Clinical Neurophysiology. Electroencephalogr Clin Neurophysiol Suppl. 1999;52:3–6.
Oostenveld R, Fries P, Maris E, Schoffelen JM. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci. 2011;2011:156869.
Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, et al. Genomewide Association Study for maximum number of alcoholic drinks in European Americans and African Americans. Alcohol Clin Exp Res. 2015;39:1137–47.
Wetherill L, Lai D, Johnson EC, Anokhin A, Bauer L, Bucholz KK, et al. Genome-wide association study identifies loci associated with liability to alcohol and drug dependence that is associated with variability in reward-related ventral striatum activity in African- and European-Americans. Genes Brain Behav. 2019;18:e12580.
Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, et al. Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav. 2019;18:e12579.
Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–82.
Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B (Methodol). 1996;58:267–88.
Knight K, Fu W. Asymptotics for lasso-type estimators. Ann Statist. 2000;28:1356–78.
Ghosh D, Chinnaiyan AM. Classification and selection of biomarkers in genomic data using LASSO. J Biomed Biotechnol. 2005;2005:147–54.
Shevade SK, Keerthi SS. A simple and efficient algorithm for gene selection using sparse logistic regression. Bioinformatics. 2003;19:2246–53.
Rijsbergen CJv. The geometry of information retrieval. Cambridge: Cambridge University Press; 2004.
Meunier D, Stamatakis EA, Tyler LK. Age-related functional reorganization, structural changes, and preserved cognition. Neurobiol Aging. 2014;35:42–54.
Charles ST, Carstensen LL. Social and emotional aging. Annu Rev Psychol. 2010;61:383–409.
Newson JJ, Thiagarajan TC. EEG frequency bands in psychiatric disorders: a review of resting state studies. Front Hum Neurosci. 2018;12:521.
van Kerkoerle T, Self MW, Dagnino B, Gariel-Mathis MA, Poort J, van der Togt C, et al. Alpha and gamma oscillations characterize feedback and feedforward processing in monkey visual cortex. Proc Natl Acad Sci USA. 2014;111:14332–41.
Rangaswamy M, Porjesz B, Chorlian DB, Choi K, Jones KA, Wang K, et al. Theta power in the EEG of alcoholics. Alcohol Clin Exp Res. 2003;27:607–15.
Affan RO, Huang S, Cruz SM, Holcomb LA, Nguyen E, Marinkovic K. High-intensity binge drinking is associated with alterations in spontaneous neural oscillations in young adults. Alcohol. 2018;70:51–60.
Lopez-Caneda E, Cadaveira F, Correas A, Crego A, Maestu F, Rodriguez Holguin S. The brain of binge drinkers at rest: alterations in theta and beta oscillations in first-year college students with a binge drinking pattern. Front Behav Neurosci. 2017;11:168.
Rangaswamy M, Porjesz B. From event-related potential to oscillations: genetic diathesis in brain (dys)function and alcohol dependence. Alcohol Res Health. 2008;31:238–42.
Park SM, Lee JY, Kim YJ, Lee JY, Jung HY, Sohn BK, et al. Neural connectivity in Internet gaming disorder and alcohol use disorder: a resting-state EEG coherence study. Sci Rep. 2017;7:1333.
Peniston EG, Kulkosky PJ. Alpha-theta brainwave training and beta-endorphin levels in alcoholics. Alcohol Clin Exp Res. 1989;13:271–9.
Kinreich S, Podlipsky I, Jamshy S, Intrator N, Hendler T. Neural dynamics necessary and sufficient for transition into pre-sleep induced by EEG neurofeedback. Neuroimage. 2014;97:19–28.
Lagopoulos J, Xu J, Rasmussen I, Vik A, Malhi GS, Eliassen CF, et al. Increased theta and alpha EEG activity during nondirective meditation. J Altern Complement Med. 2009;15:1187–92.
Menon V, Uddin LQ. Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct. 2010;214:655–67.
Xue G, Lu Z, Levin IP, Bechara A. The impact of prior risk experiences on subsequent risky decision-making: the role of the insula. Neuroimage. 2010;50:709–16.
Mies GW, Verweij KJH, Treur JL, Ligthart L, Fedko IO, Hottenga JJ, et al. Polygenic risk for alcohol consumption and its association with alcohol-related phenotypes: do stress and life satisfaction moderate these relationships? Drug Alcohol Depend. 2018;183:7–12.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators BP, VH, H Edenberg, L Bierut, includes 11 different centers: of Connecticut (V Hesselbrock); Indiana University (HJE, J Nurnberger Jr, T Foroud); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (BP); Washington University in St Louis (L Bierut, J Rice, K Bucholz, A Agrawal); University of California at San Diego (MS); Rutgers University (J Tischfield, A Brooks); University of Texas Rio Grand Valley (L Almasy), Virginia Commonwealth University (D Dick), Icahn School of Medicine at Mount Sinai (A Goate), and Howard University (R Taylor). Other COGA collaborators include: LB (University of Connecticut); J McClintick, L Wetherill, X Xuei, Y Liu, D. Lai, S O’Connor, M Plawecki, S Lourens (Indiana University); G Chan (University of Iowa; University of Connecticut); JM, DC, CK, AP, JZ (SUNY Downstate); J-CW, M Kapoor, S Bertelsen (Icahn School of Medicine at Mount Sinai); AA, V McCutcheon, S Saccone (Washington University); J Salvatore, F Aliev, B Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grand Valley). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by an NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). JLM is supported by K01DA037914 from the National Institute on Drug Abuse (NIDA), JES acknowledges support from K01AA024152 (NIAAA) and AA acknowledges support from K02DA032573 (NIDA). Funding support for GWAS genotyping performed at the Johns Hopkins University Center for Inherited Disease Research was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). GWAS genotyping was also performed at the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine, which is partially supported by NCI Cancer Center Support Grant no. P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant no. UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kinreich, S., Meyers, J.L., Maron-Katz, A. et al. Predicting risk for Alcohol Use Disorder using longitudinal data with multimodal biomarkers and family history: a machine learning study. Mol Psychiatry 26, 1133–1141 (2021). https://doi.org/10.1038/s41380-019-0534-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0534-x
This article is cited by
-
AUD-DSS: a decision support system for early detection of patients with alcohol use disorder
BMC Bioinformatics (2023)
-
Comparing two machine learning approaches in predicting lupus hospitalization using longitudinal data
Scientific Reports (2022)
-
Multimodal machine learning in precision health: A scoping review
npj Digital Medicine (2022)
-
Cleo: Smart Glasses to Monitor Consumption of Alcohol and Cigarettes
SN Computer Science (2022)
-
Alcohol dependence inpatients classification with GLM and hierarchical clustering integration using fMRI data of alcohol multiple scenario cues
Experimental Brain Research (2022)