Abstract
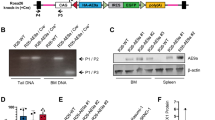
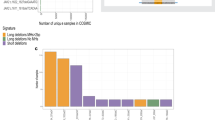
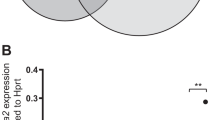
Constitutional trisomy 21 (T21) is a state of aneuploidy associated with high incidence of childhood acute myeloid leukemia (AML). T21-associated AML is preceded by transient abnormal myelopoiesis (TAM), which is triggered by truncating mutations in GATA1 generating a short GATA1 isoform (GATA1s). T21-associated AML emerges due to secondary mutations in hematopoietic clones bearing GATA1s. Since aneuploidy generally impairs cellular fitness, the paradoxically elevated risk of myeloid malignancy in T21 is not fully understood. We hypothesized that individuals with T21 bear inherent genome instability in hematopoietic lineages that promotes leukemogenic mutations driving the genesis of TAM and AML. We found that individuals with T21 show increased chromosomal copy number variations (CNVs) compared to euploid individuals, suggesting that genome instability could be underlying predisposition to TAM and AML. Acquisition of GATA1s enforces myeloid skewing and maintenance of the hematopoietic progenitor state independently of T21; however, GATA1s in T21 hematopoietic progenitor cells (HPCs) further augments genome instability. Increased dosage of the chromosome 21 (chr21) gene DYRK1A impairs homology-directed DNA repair as a mechanism of elevated mutagenesis. These results posit a model wherein inherent genome instability in T21 drives myeloid malignancy in concert with GATA1s mutations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Bulk RNA-seq data have been deposited in Gene Expression Omnibus (GSE238115) and are publicly available upon the date of publication.
References
Marlow EC, Ducore J, Kwan ML, Cheng SY, Bowles EJA, Greenlee RT, et al. Leukemia Risk in a Cohort of 3.9 Million Children with and without Down Syndrome. J Pediatr. 2021;234:172–80 e3.
Roberts I, Alford K, Hall G, Juban G, Richmond H, Norton A, et al. GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia. Blood. 2013;122:3908–17.
Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–52.
Labuhn M, Perkins K, Matzk S, Varghese L, Garnett C, Papaemmanuil E, et al. Mechanisms of progression of myeloid preleukemia to transformed myeloid leukemia in children with down syndrome. Cancer Cell. 2019;36:123–38.e10.
Laurent AP, Kotecha RS, Malinge S. Gain of chromosome 21 in hematological malignancies: lessons from studying leukemia in children with Down syndrome. Leukemia. 2020;34:1984–99.
Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–85.
Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, et al. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016;7:10754.
Knouse KA, Wu J, Amon A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 2016;26:376–84.
Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci USA. 2014;111:13409–14.
Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–46.
Biancotti JC, Narwani K, Mandefro B, Golan-Lev T, Buehler N, Hill D, et al. The in vitro survival of human monosomies and trisomies as embryonic stem cells. Stem Cell Res. 2012;9:218–24.
Schiavoni F, Zuazua-Villar P, Roumeliotis TI, Benstead-Hume G, Pardo M, Pearl FMG, et al. Aneuploidy tolerance caused by BRG1 loss allows chromosome gains and recovery of fitness. Nat Commun. 2022;13:1731.
Duijf PH, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer. 2013;132:2316–26.
Klaasen SJ, Truong MA, van Jaarsveld RH, Koprivec I, Stimac V, de Vries SG, et al. Nuclear chromosome locations dictate segregation error frequencies. Nature. 2022;607:604–9.
Camargo R, Sahoo SS, Cordoba JC, Magalhaes IQ. Germline GATA1 exon 2 mutation associated with chronic cytopenia and a non-down syndrome transient abnormal myelopoiesis with clonal trisomy 21. Leukemia. 2022;36:2347–50.
Hasle H, Kline RM, Kjeldsen E, Nik-Abdul-Rashid NF, Bhojwani D, Verboon JM, et al. Germline GATA1s-generating mutations predispose to leukemia with acquired trisomy 21 and Down syndrome-like phenotype. Blood. 2022;139:3159–65.
Maclean GA, Menne TF, Guo G, Sanchez DJ, Park IH, Daley GQ, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc Natl Acad Sci USA. 2012;109:17567–72.
Forestier E, Izraeli S, Beverloo B, Haas O, Pession A, Michalova K, et al. Cytogenetic features of acute lymphoblastic and myeloid leukemias in pediatric patients with Down syndrome: an iBFM-SG study. Blood. 2008;111:1575–83.
Byrska-Bishop M, VanDorn D, Campbell AE, Betensky M, Arca PR, Yao Y, et al. Pluripotent stem cells reveal erythroid-specific activities of the GATA1 N-terminus. J Clin Invest. 2015;125:993–1005.
Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–9.
Pfau SJ, Silberman RE, Knouse KA, Amon A. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev. 2016;30:1395–408.
Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293–9.
Wagenblast E, Araujo J, Gan OI, Cutting SK, Murison A, Krivdova G, et al. Mapping the cellular origin and early evolution of leukemia in Down syndrome. Science. 2021;373:eabf6202.
Arkoun B, Robert E, Boudia F, Mazzi S, Dufour V, Siret A, et al. Stepwise GATA1 and SMC3 mutations alter megakaryocyte differentiation in a Down syndrome leukemia model. J Clin Invest. 2022;132:e156290.
Liggett LA, Galbraith MD, Smith KP, Sullivan KD, Granrath RE, Enriquez-Estrada B, et al. Precocious clonal hematopoiesis in Down syndrome is accompanied by immune dysregulation. Blood Adv. 2021;5:1791–6.
Sullivan KD, Lewis HC, Hill AA, Pandey A, Jackson LP, Cabral JM, et al. Trisomy 21 consistently activates the interferon response. Elife. 2016;5:e16220.
Chlon TM, McNulty M, Goldenson B, Rosinski A, Crispino JD. Global transcriptome and chromatin occupancy analysis reveal the short isoform of GATA1 is deficient for erythroid specification and gene expression. Haematologica. 2015;100:575–84.
Hwang S, Cavaliere P, Li R, Zhu LJ, Dephoure N, Torres EM. Consequences of aneuploidy in human fibroblasts with trisomy 21. Proc Natl Acad Sci USA. 2021;118:e2014723118.
Guard SE, Poss ZC, Ebmeier CC, Pagratis M, Simpson H, Taatjes DJ, et al. The nuclear interactome of DYRK1A reveals a functional role in DNA damage repair. Sci Rep. 2019;9:6539.
Menon VR, Ananthapadmanabhan V, Swanson S, Saini S, Sesay F, Yakovlev V, et al. DYRK1A regulates the recruitment of 53BP1 to the sites of DNA damage in part through interaction with RNF169. Cell Cycle. 2019;18:531–51.
Malinge S, Bliss-Moreau M, Kirsammer G, Diebold L, Chlon T, Gurbuxani S, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest. 2012;122:948–62.
Sit YT, Takasaki K, An HH, Xiao Y, Hurtz C, Gearhart P, et al. Synergistic roles of DYRK1A and GATA1 in trisomy 21 megakaryopoiesis. JCI Insight. 2023;8:e172851.
Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64.
Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8.
Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54.
Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366:eaan4673.
Lambert B, Hansson K, Bui TH, Funes-Cravioto F, Lindsten J. DNA repair and frequency of x-ray and u.v.-light induced chromosome aberrations in leukocytes from patients with Down’s syndrome. Ann Hum Genet. 1976;39:293–303.
Otsuka F, Tarone RE, Seguin LR, Robbins JH. Hypersensitivity to ionizing radiation in cultured cells from Down syndrome patients. J Neurol Sci. 1985;69:103–12.
Wang Y, Chang J, Shao L, Feng W, Luo Y, Chow M, et al. Hematopoietic Stem Cells from Ts65Dn Mice Are Deficient in the Repair of DNA Double-Strand Breaks. Radiat Res. 2016;185:630–7.
Morimoto K, Kaneko T, Iijima K, Koizumi A. Proliferative kinetics and chromosome damage in trisomy 21 lymphocyte cultures exposed to gamma-rays and bleomycin. Cancer Res. 1984;44:1499–504.
Cabelof DC, Patel HV, Chen Q, van Remmen H, Matherly LH, Ge Y, et al. Mutational spectrum at GATA1 provides insights into mutagenesis and leukemogenesis in Down syndrome. Blood. 2009;114:2753–63.
Meharena HS, Marco A, Dileep V, Lockshin ER, Akatsu GY, Mullahoo J, et al. Down-syndrome-induced senescence disrupts the nuclear architecture of neural progenitors. Cell Stem Cell. 2022;29:116–30 e7.
Malinge S, Chlon T, Dore LC, Ketterling RP, Tallman MS, Paietta E, et al. Development of acute megakaryoblastic leukemia in Down syndrome is associated with sequential epigenetic changes. Blood. 2013;122:e33–43.
McGee RB, Nichols KE. Introduction to cancer genetic susceptibility syndromes. Hematology Am Soc Hematol Educ Program. 2016;2016:293–301.
Maluf SW, Erdtmann B. Genomic instability in Down syndrome and Fanconi anemia assessed by micronucleus analysis and single-cell gel electrophoresis. Cancer Genet Cytogenet. 2001;124:71–5.
Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–85.
Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. N Engl J Med. 2015;373:35–47.
Nižetić D, Groet J. Tumorigenesis in Down’s syndrome: big lessons from a small chromosome. Nat Rev Cancer. 2012;12:721–32.
Taub JW, Berman JN, Hitzler JK, Sorrell AD, Lacayo NJ, Mast K, et al. Improved outcomes for myeloid leukemia of Down syndrome: a report from the Children’s Oncology Group AAML0431 trial. Blood. 2017;129:3304–13.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
We thank the Swanson Biotechnology Center, particularly the Integrated Genomics and Bioinformatics Core, at the Koch Institute at MIT for the technical support and the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. We thank the Alana Down Syndrome Center at MIT for supporting this project. We thank the Linda Crnic Institute Human Trisome Project and Dr. Joaquin Espinosa (University of Colorado) as well as Dr. Stephanie Sherman (Emory University) for sharing the primary samples, Dr. Alan Cantor (Boston Children’s Hospital) for the CMK cell line, and Dr. Li-Huei Tsai (MIT) for the iPSC lines. We also thank Dr. John Crispino (St. Jude Children’s Research Hospital) for the scientific guidance. We are grateful for the assistance from members of the Amon, Hemann, and Rowe labs. This work was supported by the Leukemia & Lymphoma Society Career Development Program Fellowship (5500-20) and Alex’s Lemonade Stand Foundation Young Investigator Grant (22-27070) to C-CC; the St. Baldrick’s Foundation Consortium grant and Hannah’s Heroes grant to YP; the Alana Foundation grant and HHMI Investigator Award to AA; the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT, National Cancer Institute R01-CA233477, and R01-CA226898 to MTH; the National Institute of Diabetes and Digestive, and Kidney Diseases K08-DK114527, R03-DK126729, and R01-DK134515 to RGR.
Author information
Authors and Affiliations
Contributions
C-CC, RES, and AA designed the research. RES performed initial CNV experiments (4 blood samples), and C-CC performed the rest of the research (additional blood samples and iPSC experiments). DM performed CNV analysis using established HMM and CBS pipelines, and C-CC compiled the results with data from 4 samples provided by RES. JAP, DK, and YP provided experimental resources (PDX lines). RGR and MTH provided mentorship and resources following the passing of AA in 2020. C-CC wrote the manuscript. RGR and MTH reviewed and provided input on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, CC., Silberman, R.E., Ma, D. et al. Inherent genome instability underlies trisomy 21-associated myeloid malignancies. Leukemia 38, 521–529 (2024). https://doi.org/10.1038/s41375-024-02151-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-024-02151-8