Abstract
Recently, the European LeukemiaNet (ELN) revised its genetic-risk classification of acute myeloid leukemia (AML). We categorized 1637 adults with AML treated with cytarabine/anthracycline regimens according to the 2022 and 2017 ELN classifications. Compared with the 2017 ELN classification, 2022 favorable group decreased from 40% to 35% and adverse group increased from 37% to 41% of patients. The 2022 genetic-risk groups seemed to accurately reflect treatment outcomes in all patients and patients aged <60 years, but in patients aged ≥60 years, relapse rates, disease-free (DFS) and overall (OS) survival were not significantly different between intermediate and adverse groups. In younger African-American patients, DFS and OS did not differ between intermediate-risk and adverse-risk patients nor did DFS between favorable and intermediate groups. In Hispanic patients, DFS and OS did not differ between favorable and intermediate groups. Outcome prediction abilities of 2022 and 2017 ELN classifications were similar. Among favorable-risk patients, myelodysplasia-related mutations did not affect patients with CEBPAbZIP mutations or core-binding factor AML, but changed risk assignment of NPM1-mutated/FLT3-ITD-negative patients to intermediate. NPM1-mutated patients with adverse-risk cytogenetic abnormalities were closer prognostically to the intermediate than adverse group. Our analyses both confirm and challenge prognostic significance of some of the newly added markers.
Similar content being viewed by others
Introduction
Pretreatment cytogenetic findings were first to be used to prognostically stratify patients with acute myeloid leukemia (AML) [1,2,3,4,5,6,7,8,9]. Subsequently, several gene mutations were demonstrated to provide additional prognostic information [10,11,12,13,14,15,16,17,18,19]. Therefore, in 2010, the first edition of the European LeukemiaNet (ELN) recommendations for diagnosis and management of AML included a standardized system for reporting cytogenetic findings and select gene mutations to enable meaningful comparisons among studies correlating genetic findings with clinical outcome [20]. Soon thereafter, several large studies demonstrated the ability of the 2010 ELN classification to prognostically separate the favorable and adverse groups from each other and from the two intermediate groups with regard to probability of complete remission (CR) attainment, disease-free (DFS) and overall survival (OS) [21,22,23,24]. This was shown to be independent from other prognostic factors by multivariable analyses [22]. The ELN classification was then modified in 2017 by combining two intermediate groups into one, recommending mutation analysis to be performed in all patients, not only those with cytogenetically normal AML (CN-AML), considering only biallelic CEBPA mutations as prognostically favorable, requiring determination of high and low allelic ratios for internal tandem duplications of FLT3 (FLT3-ITD), and adding ASXL1, RUNX1 and TP53 mutations as adverse-risk markers [25]. The ELN genetic-risk classifications have been used in daily clinical practice to predict response to conventional chemotherapy and help guide treatment decisions, including the need for more intensive consolidation or alternative regimens.
The recently updated 2022 ELN recommendations include a revised genetic-risk classification that incorporates recent advances in our understanding of prognostic significance of genetic alterations in AML [26]. Major changes from 2017 ELN include the addition of seven myelodysplasia-related mutations to the adverse group (in the absence of favorable-risk markers); placing NPM1-mutated patients with adverse-risk cytogenetic abnormalities in the adverse group; consideration of only the presence, not allelic ratio, of FLT3-ITD; and the substitution of biallelic CEBPA mutations with in-frame mutations affecting the basic leucine zipper (bZIP) region of the CEBPA gene (CEBPAbZIP) as favorable-risk markers. Additionally, t(8;16)(p11.2;p13.3)/KAT6A::CREBBP and t(3;v)(q26.2;v)/MECOM(EVI1)-rearranged have been added to the adverse group, and hyperdiploid karyotypes with ≥3 trisomies without structural abnormality are no longer considered as complex [26].
The goals of our study were to assess how well the 2022 ELN genetic-risk groups associate with treatment response, DFS and OS, to compare the performance of the 2022 and 2017 ELN classifications both in all patients and in age cohorts, and to assess the effectiveness of newly introduced features in outcome prediction.
Methods
Patients and treatment
We analyzed 1637 adults diagnosed with de novo AML (other than acute promyelocytic leukemia, which is not included in the 2022 ELN guidelines), including 1040 patients aged <60 years (hereafter referred to as younger) and 597 patients aged ≥60 years (older), who were treated on Cancer and Leukemia Group B (CALGB) frontline treatment protocols between 1986 and 2013. CALGB is now part of the Alliance for Clinical Trials in Oncology (Alliance). For definition of race and ethnicity please see Supplementary Information. Younger patients received intensive cytarabine/daunorubicin-based induction chemotherapy and consolidation with high-dose chemotherapy or autologous hematopoietic stem-cell transplantation (HSCT; details of treatment trials are provided in the Supplementary Information). All patients aged ≥60 years received cytarabine/daunorubicin-based chemotherapy.
Analyses of DFS and OS were conducted on patients who did not receive an allogeneic HSCT in first CR (94% of patients; 6% underwent HSCT). All patients provided written informed consent to participate in treatment studies and for the research use of their specimens before enrollment in agreement with the Declaration of Helsinki. Study protocols were approved by the Institutional Review Board at each center.
Cytogenetic and molecular genetic analyses
Cytogenetic analyses of pretreatment bone marrow (BM) and/or blood samples were performed by CALGB/Alliance-approved institutional laboratories and the results confirmed by central karyotype review [27]. CN-AML was determined by analysis of ≥20 metaphase cells from BM subjected to short-term (24–48-h) unstimulated cultures [27].
The mutational status of the ASXL1, BCOR, EZH2, NPM1, RUNX1, SF3B1, SRSF2, STAG2, TP53, U2AF1 and ZRSR2 genes was determined centrally at The Ohio State University in patients’ DNA extracted from viably frozen cells collected via companion protocol CALGB 20202 by targeted amplicon sequencing using the MiSeq platform (Illumina, San Diego, CA) [28]. Testing for FLT3-ITD was done using the Sanger sequencing method [13]. Detection of CEBPAbZIP mutations was performed according to the 2022 ELN guidelines [26] using mutational profiling via targeted amplicon sequencing and/or transcriptional profiling [29]. Further experimental details are provided in the Supplementary Information.
Clinical endpoints and statistical analysis
Clinical endpoints were defined according to generally accepted criteria [20, 25, 26, 30] and treatment study protocols.
DFS was measured from the date of CR until the date of relapse or death from any cause, and relapse-free patients were censored at the last follow-up. OS was measured from the date on study until the date of death, and patients alive at last follow-up were censored. Data quality was ensured by review of data by the Alliance Statistics and Data Management Center and by the chairpersons of included studies following Alliance policies.
Pretreatment characteristics were compared using the Fisher’s exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively. For time-to-event analyses, we calculated survival estimates using the Kaplan-Meier method and compared groups using the log-rank test [31]. For comparisons of predictive values of the 2022 and 2017 ELN genetic-risk classifications, we used receiver operating characteristic (ROC) curves as graphical plots. The areas under the curve (AUC) are provided together with the 95% confidence intervals. An AUC = 0.50 denotes lack of prediction ability, equivalent to that of random chance or a coin flip, whereas an AUC = 1.00 (highest possible value) indicates perfect prediction ability. Commonly accepted criteria are that an AUC of 0.6–0.69, 0.7–0.79, 0.8–0.89, and ≥0.9 indicate, respectively, poor, fair, good and very good prediction ability [32, 33]. In our models with a binary outcome, the AUC is equal to the more commonly known c-statistic [31,32,33,34]. The binary outcomes we considered were CR achievement (yes/no), relapse (yes/no), DFS (relapsed or dead versus relapse-free at three years) and OS (dead or alive at three years).
The dataset was locked on August 24, 2022. Data collection and statistical analyses were performed by the Alliance Statistics and Data Management Center using SAS 9.4 and TIBCO Spotfire S + 8.2. The median follow-up for patients still alive was 7.6 years.
Results
The distribution of risk-groups according to the 2022 and 2017 ELN classifications
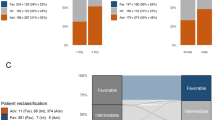
Per the 2022 ELN classification, 35% of patients were assigned to the favorable group, 24% to intermediate and 41% to the adverse group. This represents a decrease in the proportion of patients in the favorable group and increase in the proportion of patients in the adverse group compared with the respective genetic-risk groups using 2017 ELN classification, namely, 40%, 23%, and 37% (Fig. 1a). More detailed reallocation of patients from 2017 ELN genetic-risk groups into the 2022 ELN ones is illustrated by Supplementary Fig. 1.
a Bar charts depicting the distribution of the genetic-risk groups in all patients categorized according to the 2022 ELN classification and of those grouped according to the 2017 ELN guidelines. b Disease-free survival and c overall survival of all patients classified according to the 2022 ELN guidelines.
Associations between 2022 ELN genetic-risk groups and clinical outcome
Eighty-four percent of favorable-risk patients achieved a CR, compared with 68% of intermediate-risk and 44% of adverse-risk patients (P < 0.001, Table 1). Relapse rates also differed among genetic-risk groups (52% vs 72% vs 87%, P < 0.001) as did DFS (5-year rates, 44% vs 21% vs 6%, P < 0.001; Fig. 1b) and OS (5-year rates, 48% vs 22% vs 7%, P < 0.001; Fig. 1c; Table 1). Multivariable analyses revealed that CR rates, DFS and OS remained better for patients in the favorable and intermediate groups compared with patients in the adverse group (P < 0.001 for all comparisons) after adjustment for such established prognostic factors in AML as age, WBC or platelets (Supplementary Table 1). Thus, the 2022 ELN genetic-risk groups seemed to accurately reflect patient outcomes with respect to achievement of CR, relapse rates, DFS and OS. However, when we compared the abilities of outcome prediction between the 2022 and 2017 ELN classifications, we found no significant differences with regard to CR achievement [area under the curve (AUC), 0.70 vs 0.71, P = 0.44], relapse rates (AUC, 0.67 vs 0.67, P = 0.74); DFS (AUC, 0.71 vs 0.70, P = 0.74) and OS (AUC, 0.74 vs 0.73, P = 0.15; Supplementary Fig. 2a–d), indicating that the 2022 ELN genetic-risk classification does not represent a clinically relevant increment of improvement over the 2017 ELN classification for outcome prediction in our adult patients with AML aged 17–89 years.
Breakdown of genetic-risk groups in younger and older patients for 2022 and 2017 ELN classifications and associations with outcome
It is well established that the distributions of many cytogenetic abnormalities and gene mutations differ between younger and older patients [19, 22, 28, 35,36,37,38,39,40], as did the distributions of genetic-risk groups and their associations with outcome in 2010 ELN [21, 22] and 2017 ELN [41,42,43] classifications. Moreover, the availability of new regimens for older and/or unfit patients makes assessment of the likelihood to respond to therapy separately in older and younger patients of importance. Thus, we assessed the risk-group distributions and outcomes of patients aged <60 years and those aged ≥60 years. As Figs. 2a and 3a illustrate, younger patients were more often classified in the favorable (42% vs 24%) and intermediate (27% vs 19%) 2022 ELN risk-groups than older patients, whereas the latter were more frequent in the adverse (58% vs 31%) risk-group. Compared with the 2017 ELN classification, the proportions of younger patients in the 2022 ELN favorable group decreased (42% vs 47%) and in intermediate group increased (27% vs 23%). Among older patients, the 2022 ELN criteria resulted in smaller favorable (24% vs 30%) and intermediate (19% vs 22%) groups and an enlarged adverse group (58% vs 48%), compared with the 2017 ELN classification.
a Bar charts depicting the distribution of the genetic-risk groups in younger patients categorized according to the 2022 ELN classification and of those grouped according to the 2017 ELN guidelines. b Disease-free survival and c overall survival of younger patients classified according to the 2022 ELN guidelines.
a Bar charts depicting the distribution of the genetic-risk groups in older patients categorized according to the 2022 ELN classification and of those grouped according to the 2017 ELN guidelines. b Disease-free survival and c overall survival of older patients classified according to the 2022 ELN guidelines.
Concerning treatment outcomes of younger patients, the 2022 ELN genetic-risk groups essentially associated with the expected outcomes, with the attainment of CR, relapse rates, DFS and OS differing significantly among the favorable, intermediate and adverse groups (Fig. 2b, c; Supplementary Table 2). Among patients aged ≥60 years, those in the favorable group had better outcome than patients in both remaining groups. However, with the exception of CR rates, which were higher for intermediate- than adverse-risk patients (54% vs 35% P < 0.001), the outcome of patients classified in the intermediate group was very poor and did not differ significantly from outcome of the adverse group with regard to relapse rates (89% vs 88% P = 1.00), DFS (5-year rates, 5% vs 5%, P = 0.27) or OS (5-year rates, 6% vs 2%, P = 0.09; Fig. 3b, c, Supplementary Table 3).
We also performed exploratory analyses of the outcomes of younger patients of African-American ancestry and those self-identifying as Hispanics. We found no significant differences in DFS of African-American patients between the 2022 ELN favorable and intermediate (5-year rates, 32% vs 30%, P = 0.42) groups nor in DFS (5-year rates, 30% vs 0%, P = 0.30) and OS (5-year rates, 24% vs 3%, P = 0.46) between intermediate and adverse groups (Supplementary Fig. 3a, b, Supplementary Table 4). Moreover, among younger Hispanic patients, we observed no significant differences in DFS (5-year rates, 47% vs 67%, P = 0.42) or OS (5-year rates, 61% vs 71%, P = 0.67) between the 2022 ELN favorable and intermediate groups (Supplementary Fig. 3c, d, Supplementary Table 5). There were not enough patients aged ≥60 years in either racial-ethnic group for similar analyses.
Next, we compared the abilities of outcome prediction between the 2022 ELN and 2017 ELN classifications separately in younger and older patients. We found no significant advantage for the use of 2022 ELN classification over the 2017 ELN one in either the younger (Supplementary Fig. 4a–d) or older patients (Supplementary Fig. 5a–d).
Assessment of the newly introduced prognostic markers
AML with myelodysplasia-related gene mutations
A major change in the 2022 ELN classification was the addition of seven myelodysplasia-related gene mutations (in the BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1 and ZRSR2 genes) to the ASXL1, RUNX1 and TP53 mutations already included in the 2017 ELN classification as criteria for adverse group assignment, unless they co-exist with “favorable-risk AML subtypes” [26]. Indeed, in our cohort, patients with myelodysplasia-related mutations and no favorable genetic features had a low CR rate similar to the CR rate of patients with other adverse-risk markers (43% vs 44%, P = 0.75) and their DFS was not significantly different (5-year rates, 5% vs 8%, P = 0.10). However, patients with myelodysplasia-related mutations had longer OS (1-year rates, 38% vs 24%, 5-year rates, 7% vs 7%, P = 0.005). Conversely, the outcome of patients with myelodysplasia-related mutations without favorable-risk features was worse than the outcome of patients harboring myelodysplasia-related mutations together with favorable genetic-risk markers (CR rates, 43% vs 73%, P < 0.001; DFS, 5-year rates, 5% vs 39%, P < 0.001; OS, 5-year rates, 7% vs 39%, P < 0.001; Fig. 4a, b, Supplementary Table 6).
a Disease-free survival and b overall survival of patients with myelodysplasia-related mutations with and those without favorable-risk AML subtypes, and of patients in the 2022 ELN adverse group who do not harbor myelodysplasia-related mutations. c Disease-free survival and d overall survival of patients in the 2022 ELN favorable group with myelodysplasia-related mutations and favorable-risk AML subtypes, and of patients in the 2022 ELN favorable group who do not have myelodysplasia-related mutations. e Disease-free survival and f overall survival of patients in the 2022 ELN favorable group with NPM1 mutations, no FLT3-ITD and myelodysplasia-related mutations, and of patients in the 2022 ELN favorable group with NPM1 mutations without FLT3-ITD or myelodysplasia-related mutations.
However, the outcome of patients with favorable-risk AML harboring myelodysplasia-related mutations was worse than the outcome of patients with favorable-risk AML without myelodysplasia-related mutations (CR rates, 73% vs 86%, P = 0.004; OS, 5-year rates, 39% vs 50%, P = 0.003), although the difference in DFS was not statistically significant (5-year rates, 39% vs 45%, P = 0.14; Fig. 4c, d, Supplementary Table 7).
Notably, the favorable genetic-risk group comprises three distinct subtypes, namely NPM1-mutated patients without FLT3-ITD, patients with core-binding factor AML (CBF-AML), and those with CEBPAbZIP mutations. Thus, we next tested whether co-occurring myelodysplasia-related mutations impacted each of the aforementioned subsets alike. We found that co-occurring myelodysplasia-related mutations did not substantially affect the favorable impact of CBF-AML (CBF-AML with myelodysplasia-associated mutations vs CBF-AML without: CR rates, 93% vs 92%, P = 1.00; DFS, 5-year rates, 57% vs 51%, P = 0.65; OS, 5-year rates, 66% vs 63%, P = 0.65; Supplementary Table 8, Supplementary Fig. 6a, b), or of CEBPAbZIP mutations (CR rates: 76% vs 85%, P = 0.33; DFS, 5-year rates, 38% vs 43%, P = 0.74; OS, 5-year rates: 38% vs 50%, P = 0.49; Supplementary Table 9, Supplementary Fig. 6c, d). However, the presence of myelodysplasia-related mutations resulted in worsening treatment outcome of NPM1-mutated/FLT3-ITD-negative patients, who had lower CR rates (67% vs 81%, P = 0.02), and shorter DFS (5-year rates, 30% vs 43%, P = 0.03) and OS (5-year rates, 32% vs 42%, P = 0.005) than NPM1-mutated/FLT3-ITD-negative patients without myelodysplasia-associated mutations (Fig. 4e, f, Supplementary Table 10). Consequently, we compared outcome of the NPM1-mutated/FLT3-ITD-negative patients who carried myelodysplasia-related mutations with outcome of patients included in the 2022 ELN intermediate group. Surprisingly, we found no significant differences between these subsets in CR rates (67% vs 68%, P = 1.00), relapse rates (70% vs 72%, P = 0.71), DFS (5-year rates, 30% vs 21%, P = 0.19) or OS (5-year rates, 32% vs 22%, P = 0.28; Supplementary Table 11). These results suggest that the prognostic significance of myelodysplasia-related mutations that “co-occur with favorable-risk AML subtypes” is not the same for all genetic subtypes comprising the 2022 ELN favorable group.
AML with NPM1 mutations co-occurring with adverse-risk cytogenetic features
Another refinement of the 2022 ELN genetic-risk classification was that the presence of adverse-risk cytogenetic abnormalities in NPM1-mutated AML now defines adverse risk [26]. Hence, we compared the outcome of NPM1-mutated patients classified in the favorable group (i.e., NPM1-mutated patients without FLT3-ITD) with outcome of NPM1-mutated/FLT3-ITD-negative patients with adverse-risk chromosome abnormalities. We found that although the CR rate of NPM1-mutated patients with adverse-risk abnormalities was lower than that of patients without adverse-risk abnormalities, the difference was not significant (64% vs 79%, P = 0.20), nor was the difference in relapse rates (88% vs 56%, P = 0.14). However, OS of NPM1-mutated patients with adverse-risk abnormalities was shorter (5-year rates, 23% vs 41%, P = 0.04) and DFS tended to be worse (5-year rates, 38% vs 41%, P = 0.06; Supplementary Table 12) than those of patients without adverse-risk cytogenetics. We then compared the former subset with other patients in the 2022 ELN adverse group. Although there were no significant differences for any of the endpoints analyzed, CR rates (P = 0.17), DFS (P = 0.12) and OS (P = 0.08), they all tended to be better for NPM1-mutated patients with adverse-risk abnormalities than for other adverse-risk patients (Supplementary Table 12). This prompted us to check whether the outcomes of NPM1-mutated patients with adverse-risk abnormalities were different from or similar to outcome of patients in the 2022 ELN intermediate group. As data in the Fig. 5a, b and Supplementary Table 13 show, NPM1-mutated patients with adverse-risk abnormalities were much closer prognostically to the 2022 ELN intermediate than adverse group.
a Disease-free survival and b overall survival of patients with NPM1 mutations and no FLT3-ITD categorized according to the presence or absence of adverse-risk cytogenetic features. Outcomes of patients classified in the 2022 ELN favorable and adverse (excluding NPM1-mutated patients with adverse-risk cytogenetic features) groups are shown for comparison. c Disease-free survival and d overall survival of intermediate-risk patients who harbor FLT3-ITD, compared with other patients included in the intermediate group and of patients in the adverse group.
Additionally, the 2022 ELN guidelines classify NPM1-mutated patients who harbor FLT3-ITD in the intermediate group, unless they also have adverse cytogenetics, which re-assigns such patients to the adverse group. Unfortunately, there were only four patients with NPM1 mutations, FLT3-ITD and adverse cytogenetic abnormalities in our study, which precluded further analysis.
AML with FLT3-internal tandem duplication
AML with FLT3-ITD is now categorized in the intermediate group, irrespective of the allelic ratio or concurrent presence of NPM1 mutations [26]. As this change was partially justified by the modifying impact of midostaurin-based therapy on FLT3-ITD without NPM1 mutation, we separately analyzed the survival of patients treated with chemotherapy only and of those receiving midostaurin on the CALGB 10603 (RATIFY) protocol. Among patients receiving chemotherapy, those with FLT3-ITD had worse relapse rate (P = 0.01), DFS (P < 0.001) and OS (P < 0.001), and lower CR rates, but not significantly so (P = 0.08), than other patients included in the 2022 ELN intermediate group. Compared with the adverse group, although FLT3-ITD-positive patients had higher CR rates (P < 0.001) and longer OS (P < 0.001), their relapse rates (P = 0.21) and DFS (P = 0.34) were not significantly different (Fig. 5c, d, Supplementary Table 14). There were no significant differences in CR (P = 0.82) or relapse rates (P = 1.00), DFS (P = 0.14) or OS (P = 0.11) between a relatively small cohort of patients with FLT3-ITD treated with midostaurin and those receiving chemotherapy only (Supplementary Table 15, Supplementary Fig. 7a, b).
AML with CEBPAbZIP mutations
One of the notable changes to the criteria used for classifying patients into the 2022 ELN favorable genetic-risk group was replacing biallelic CEBPA mutations by the in-frame CEBPAbZIP mutations [26]. To assess this change, we first prepared Kaplan-Meier curves illustrating outcomes of all patients with biallelic CEBPA mutations (regardless whether they included CEBPAbZIP mutations or not) and of all patients with CEBPAbZIP mutations (irrespective whether these were monoallelic or biallelic mutations; Supplementary Fig. 8a, b). We could not formally compare these patient groups because some patients were included in both groups. Nevertheless, although OS of patients with CEBPAbZIP mutations seemed slightly better, the improvement over biallelic CEBPA mutations did not appear compelling and the Kaplan-Meier curves representing DFS overlapped. We then compared outcomes of three subsets among CEBPA-mutated patients: those with monoallelic CEBPAbZIP mutations, with biallelic CEBPAbZIP mutations and with biallelic non-bZIP CEBPA mutations. As expected, patients in the last aforementioned group had the worst outcome. Somewhat surprisingly, patients with biallelic CEBPAbZIP mutations had higher CR rates (95% vs 64%, P < 0.001), and longer OS (5-year rates, 53% vs 38%, P = 0.009), but not DFS (P = 0.27), than patients with monoallelic CEBPAbZIP mutations (Supplementary Fig. 8c, d; Supplementary Table 16).
Discussion
In this study of a large patient cohort with long follow-up, we applied the revised 2022 ELN criteria to stratify patients with de novo AML into genetic-risk groups and compared the performance of the modified classification with the previous one published in 2017. In the entire cohort, patients assigned to the 2022 ELN favorable group had better outcomes than those in the intermediate group, whose outcome was better than outcome of patients in the adverse group. Using ROC curves and the area under the curve we found that according to commonly accepted criteria [32, 33] the prediction ability of the 2022 ELN classification was fair for attainment of CR, DFS and OS, but poor for predicting relapse. Importantly, despite newly introduced modifications, the predictive ability of the 2022 ELN genetic-risk classification was essentially the same as predictive ability of the 2017 ELN classification with regard to all outcome endpoints tested.
Since the distribution of ELN genetic-groups differs between younger and older adults, with the former being more often classified in the favorable and the latter in the adverse groups, we analyzed patients aged <60 years and those aged ≥60 years separately. While in the younger patients the 2022 ELN classification separated genetic-risk groups quite well, among older adults only those in the favorable group had better outcomes, whereas there were no significant differences in relapse rates, DFS or OS between the intermediate and adverse groups. In a previous study, we observed a similar phenomenon in older patients classified according to the 2017 ELN criteria [42]. Therefore, our data support previous suggestions [22] that the ELN genetic-risk classification should be tailored to younger and older adults separately, which is also supported by the availability of treatment options targeting specific genetic alterations [44,45,46,47,48], whose incidence differs between younger and older patients [19, 22, 28, 35,36,37,38,39,40,41,42,43].
Moreover, our preliminary results indicate the need for large studies focused on racial/ethnic groups such as African-American and Hispanic patients. We found no significant differences in survival between 2022 ELN favorable and intermediate and between intermediate and adverse groups in patients of African-American ancestry, and no significant difference in DFS between favorable and intermediate groups in Hispanic patients. Although these results may be in part related to the relatively low number of patients we were able to analyze, previously identified racial/ethnic differences in the distribution of genetic alterations and outcomes [49,50,51,52,53] warrant application of 2022 ELN criteria to larger cohorts of African-American and Hispanic patients to confirm or refute our observations.
Among major aims of our study was evaluation of the new markers used for 2022 ELN group assignment. First, we found that patients harboring one or more myelodysplasia-related gene mutations without favorable-risk features had indeed very poor outcome placing them in the adverse group, and that the co-existence of favorable-risk features substantially improved patient outcomes. However, this improvement has not been sufficient to place the entire cohort of patients with favorable-risk AML and myelodysplasia-related mutations in the 2022 ELN favorable group, since their CR rates and OS were significantly worse. Importantly, the 2022 ELN favorable group is not homogeneous and consists of three major subsets. Our analyses revealed that myelodysplasia-related mutations did not negatively affect outcome of patients with CBF-AML and those harboring CEBPAbZIP mutations, who would still be classified in the favorable group. However, the presence of myelodysplasia-related mutations in NPM1-mutated/FLT3-ITD-negative patients portended worse outcome, which placed these patients firmly in the 2022 ELN intermediate, not favorable, group. If these results are corroborated, future editions of the ELN classification should consider modification of risk-assignment for the aforementioned patient subsets.
Likewise, we have confirmed that patients with NPM1 mutations co-occurring with adverse-risk cytogenetic features have worse prognosis than other NPM1-mutated patients included in the 2022 ELN favorable group. However, in contrast to the results of a recent study [54], the former subset seems to be closer prognostically to the 2022 ELN intermediate rather than adverse group. Unfortunately, the low number of patients with NPM1 mutations and adverse-risk cytogenetics (n = 14) precludes us from making a definitive recommendation and shows the need for further study. Likewise, it is also necessary to confirm in larger patient cohorts that patients with FLT3-ITD belong to the intermediate group.
Based on recent reports [29, 55, 56], 2022 ELN recommendations replaced biallelic CEBPA mutations with the in-frame CEBPAbZIP mutations as a criterion for classifying patients into the favorable genetic-risk group. We have generally confirmed that CEBPAbZIP mutations confer better prognosis than biallelic, non-bZIP CEBPA mutations. Rather surprisingly, however, we found that patients with biallelic CEBPAbZIP mutations had higher CR rates and longer OS than patients with monoallelic CEBPAbZIP mutations, which differs from previously reported data [29, 55, 56]. The reasons for this discrepancy are unclear, but may be partially attributed to differences in treatment administered in different studies.
Among limitations of our study is its retrospective nature and the inability to assess the outcomes of patients with cytogenetic markers newly added to the 2022 ELN genetic-risk classification such as prognostically adverse t(8;16)(p11.2;p13.3)/KAT6A::CREBBP and t(3;v)(q26.2;v)/MECOM(EVI1)-rearranged, or hyperdiploid complex karyotypes with three or more trisomies without structural abnormalities that are no longer considered to be adverse markers. All these chromosome abnormalities occurred in too few patients for meaningful analyses in our study, thus warranting further collaborative efforts involving large AML study groups.
In summary, our large study assessing the newly revised 2022 ELN genetic-risk classification confirms its usefulness for prognostic stratification of patients with de novo AML. However, despite introduction of several new criteria, we found no advantage of this classification over the previous one published in 2017. Importantly, our data support earlier calls for separating younger from older adults because both the incidence of genetic alterations and outcomes differ between these age groups [19, 22, 28, 35,36,37,38,39,40,41,42,43]. It is hoped that future ELN recommendations will consider our and other [38, 57, 58] suggestions for refinement of the genetic-risk classification of AML.
Data availability
Patient data used in survival analyses were obtained from the Alliance Statistics and Data Management Center. Individual participant data will not be shared.
References
Arthur DC, Berger R, Golomb HM, Swansbury GJ, Reeves BR, Alimena G, et al. The clinical significance of karyotype in acute myelogenous leukemia. Cancer Genet Cytogenet. 1989;40:203–16.
Mrózek K, Heinonen K, de la Chapelle A, Bloomfield CD. Clinical significance of cytogenetics in acute myeloid leukemia. Semin Oncol. 1997;24:17–31.
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–33.
Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood. 2000;96:4075–83.
Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–36.
Fröhling S, Schlenk RF, Kayser S, Morhardt M, Benner A, Döhner K, et al. Cytogenetics and age are major determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: results from AMLSG trial AML HD98-B. Blood. 2006;108:3280–8.
Farag SS, Archer KJ, Mrózek K, Ruppert AS, Carroll AJ, Vardiman JW, et al. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: results from Cancer and Leukemia Group B 8461. Blood. 2006;108:63–73.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65.
Mrózek K. Molecular cytogenetics in acute myeloid leukemia in adult patients: practical implications. Pol Arch Intern Med. 2022;132:16300.
Fröhling S, Schlenk RF, Stolze I, Bihlmayr J, Benner A, Kreitmeier S, et al. CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutations. J Clin Oncol. 2004;22:624–33.
Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–87.
Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CAJ, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117:2469–75.
Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–9.
Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–35.
Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66.
Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–6.
Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood. 2006;107:4011–20.
Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–48.
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604.
Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–74.
Röllig C, Bornhäuser M, Thiede C, Taube F, Kramer M, Mohr B, et al. Long-term prognosis of acute myeloid leukemia according to the new genetic risk classification of the European LeukemiaNet recommendations: evaluation of the proposed reporting system. J Clin Oncol. 2011;29:2758–65.
Mrózek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–23.
Mrózek K, Nicolet D, Maharry KS, Carroll AJ, Marcucci G, Bloomfield CD. Reply to K. Orendi et al. [letter]. J Clin Oncol. 2013;31:2361–2.
Alpermann T, Kern W, Schnittger S, Schmid C, Kreuzer KA, Serve H, et al. Evaluation of the proposed reporting system of the European LeukemiaNet and recommendations for prognosis of acute myeloid leukemia. Leuk Res. 2013;37:197–200.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Mrózek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–44.
Eisfeld A-K, Mrózek K, Kohlschmidt J, Nicolet D, Orwick S, Walker CJ, et al. The mutational oncoprint of recurrent cytogenetic abnormalities in adult patients with de novo acute myeloid leukemia. Leukemia. 2017;31:2211–8.
Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Röllig C, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139:87–103.
Cheson BD, Cassileth PA, Head DR, Schiffer CA, Bennett JM, Bloomfield CD, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–9.
Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE Regression methods in biostatistics: linear, logistic, survival and repeated measures models. New York, NY, USA: Springer; 2005.
Walter RB, Othus M, Burnett AK, Löwenberg B, Kantarjian HM, Ossenkoppele GJ, et al. Resistance prediction in AML: analysis of 4601 patients from MRC/NCRI, HOVON/SAKK, SWOG and MD Anderson Cancer Center. Leukemia. 2015;29:312–20.
Walter RB, Othus M, Borthakur G, Ravandi F, Cortes JE, Pierce SA, et al. Prediction of early death after induction therapy for newly diagnosed acute myeloid leukemia with pretreatment risk scores: a novel paradigm for treatment assignment. J Clin Oncol. 2011;29:4417–23.
Godwin CD, Othus M, Powell MA, Buckley SA, Estey EH, Walter RB. Prediction of early death in adults with relapsed or refractory acute myeloid leukemia [letter]. Leuk Lymphoma. 2016;57:2421–4.
Schoch C, Schnittger S, Kern W, Dugas M, Hiddemann W, Haferlach T. Acute myeloid leukemia with recurring chromosome abnormalities as defined by the WHO-classification: incidence of subgroups, additional genetic abnormalities, FAB subtypes and age distribution in an unselected series of 1,897 patients with acute myeloid leukemia [letter]. Haematologica. 2003;88:351–2.
Moorman AV, Roman E, Willett EV, Dovey GJ, Cartwright RA, Morgan GJ. Karyotype and age in acute myeloid leukemia. Are they linked? Cancer Genet Cytogenet. 2001;126:155–61.
Mrózek K. Cytogenetic, molecular genetic, and clinical characteristics of acute myeloid leukemia with a complex karyotype. Semin Oncol. 2008;35:365–77.
Ostronoff F, Othus M, Lazenby M, Estey E, Appelbaum FR, Evans A, et al. Prognostic significance of NPM1 mutations in the absence of FLT3-internal tandem duplication in older patients with acute myeloid leukemia: a SWOG and UK National Cancer Research Institute/Medical Research Council report. J Clin Oncol. 2015;33:1157–64.
Straube J, Ling VY, Hill GR, Lane SW. The impact of age, NPM1mut, and FLT3ITD allelic ratio in patients with acute myeloid leukemia [letter]. Blood. 2018;131:1148–53.
Grimwade D, Mrózek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am. 2011;25:1135–61. vii
Herold T, Rothenberg-Thurley M, Grunwald VV, Janke H, Goerlich D, Sauerland MC, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34:3161–72.
Eisfeld A-K, Kohlschmidt J, Mrózek K, Blachly JS, Walker CJ, Nicolet D, et al. Mutation patterns identify adult patients with de novo acute myeloid leukemia aged 60 years or older who respond favorably to standard chemotherapy: an analysis of Alliance studies. Leukemia. 2018;32:1338–48.
Eisfeld A-K, Kohlschmidt J, Mims A, Nicolet D, Walker CJ, Blachly JS, et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years. Leukemia. 2020;34:3215–27.
Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377:454–64.
Marcucci G, Geyer S, Laumann K, Zhao W, Bucci D, Uy GL, et al. Combination of dasatinib with chemotherapy in previously untreated core binding factor acute myeloid leukemia: CALGB 10801. Blood Adv. 2020;4:696–705.
Montesinos P, Recher C, Vives S, Zarzycka E, Wang J, Bertani G, et al. Ivosidenib and azacitidine in IDH1-mutated acute myeloid leukemia. N Engl J Med. 2022;386:1519–31.
Walker AR, Byrd JC, Blachly JS, Bhatnagar B, Mims AS, Orwick S, et al. Entospletinib in combination with induction chemotherapy in previously untreated acute myeloid leukemia: response and predictive significance of HOXA9 and MEIS1 expression. Clin Cancer Res. 2020;26:5852–9.
Burd A, Levine RL, Ruppert AS, Mims AS, Borate U, Stein EM, et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the Beat AML Master Trial. Nat Med. 2020;26:1852–8.
Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–17.
Bierenbaum J, Davidoff AJ, Ning Y, Tidwell ML, Gojo I, Baer MR. Racial differences in presentation, referral and treatment patterns and survival in adult patients with acute myeloid leukemia: a single-institution experience. Leuk Res. 2012;36:140–5.
Bhatnagar B, Kohlschmidt J, Mrózek K, Zhao Q, Fisher JL, Nicolet D, et al. Poor survival and differential impact of genetic features of Black patients with acute myeloid leukemia. Cancer Discov. 2021;11:626–37.
Bhatnagar B, Eisfeld A-K. Racial and ethnic survival disparities in patients with haematological malignancies in the USA: time to stop ignoring the numbers. Lancet Haematol. 2021;8:e947–e54.
Larkin KT, Nicolet D, Kelly BJ, Mrózek K, LaHaye S, Miller KE, et al. High early death rates, treatment resistance, and short survival of Black adolescents and young adults with AML. Blood Adv. 2022;6:5570–81.
Angenendt L, Röllig C, Montesinos P, Ravandi F, Juliusson G, Récher C, et al. Revisiting co-existing chromosomal abnormalities in NPM1-mutated AML in light of the revised ELN 2022 classification [letter]. Blood. 2023;141:433–5.
Tarlock K, Lamble AJ, Wang Y-C, Gerbing RB, Ries RE, Loken MR, et al. CEBPA-bZip mutations are associated with favorable prognosis in de novo AML: a report from the Children’s Oncology Group [published correction appears in Blood. 2022;139:1601]. Blood. 2021;138:1137–47.
Wakita S, Sakaguchi M, Oh I, Kako S, Toya T, Najima Y, et al. Prognostic impact of CEBPA bZIP domain mutation in acute myeloid leukemia. Blood Adv. 2022;6:238–47.
Tsai C-H, Yao C-Y, Tien F-M, Tang J-L, Kuo Y-Y, Chiu Y-C, et al. Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine. 2019;40:240–50.
Pogosova-Agadjanyan EL, Moseley A, Othus M, Appelbaum FR, Chauncey TR, Chen IL, et al. AML risk stratification models utilizing ELN-2017 guidelines and additional prognostic factors: a SWOG report. Biomark Res. 2020;8:29.
Acknowledgements
Celebrating the life and accomplishments of Dr. Clara D. Bloomfield (1942–2020), who died unexpectedly on March 1, 2020. The authors are grateful to the patients who consented to participate in these clinical trials and the families who supported them; to Christopher Manring and the CALGB/Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services; and to Lisa J. Sterling for data management. Research reported in this publication was supported by an allocation of computing resources from The Ohio Supercomputer Center and Shared Resources (Leukemia Tissue Bank). Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology), UG1CA189824, UG1CA233338, UG1CA233339, P50 CA140158, UG1CA233331, R35CA197734, UG1CA189850, UG1CA233180, UG1CA233327, and 5P30CA016058; the Coleman Leukemia Research Foundation; ASH Junior Faculty Scholar and ASH Bridge Award (A-KE); the Leukemia & Lymphoma Society Translational Research Grant (A-KE); The Leukemia Research Foundation (A-KE); the National Comprehensive Cancer Network Foundation Young Investigator Award (JSB); the Alliance for Clinical Trials in Oncology Scholar Award (JSB); The D Warren Brown Foundation; Pelotonia (A-KE, ASM). Support to Alliance for Clinical Trials in Oncology and Alliance Foundation Trials programs is listed at https://acknowledgments.alliancefound.org. Trial Registration Numbers are NCT00048958, NCT00899223, NCT00900224. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Contribution: KM, JK and A-KE designed the study; KM, JK, KJA, KTL, CCO, JCB and A-KE contributed to the data interpretation; KM, JK, and A-KE wrote the manuscript; A-KE, ASM, JSB, and SO performed laboratory-based research and genomic data analysis; JK and DN performed statistical analysis; AJC, KM, JEK, BLP, WGB, GM, MRB, GLU, WS and JCB were involved directly or indirectly in the care of patients and/or sample procurement. All authors read and agreed on the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
JSB is a consultant for and reported honoraria from KITE, INNATE, AstraZeneca and AbbVie. ASM received research funding from Leukemia and Lymphoma Society’s Beat AML clinical study, Aptevo, Daiichi Sankyo, Glycomemetics, Kartos Pharmaceuticals, Xencor and Genentech; and is a consultant for Aptevo, Abbvie, BMS, Glycomemetics, Jazz Pharmaceuticals, Kura Oncology, and Syndax Pharmaceuticals. BLP is a consultant for Cornerstone Pharmaceuticals and reported research funding from Ambit Biosciences, Cornerstone, Genentech, Hoffman LaRoche, Jazz Pharmaceuticals, Novartis and Pfizer. WGB reported honoraria from Abbvie, Syndax, and AmerisourceBergen and research funding from Celyad Oncology, Nkarta, Xencor, Forma Therapeutics and Leukemia and Lymphoma Society. GLU is a consultant for AbbVie, Agios, Jazz, GlaxoSmithKline, Genentech, and Novartis; reported honoraria from Astellas and research funding from Macrogenics. JCB consults for Astellas, AstraZeneca, Novartis, Pharmacyclics, Syndax and Trillium; receives honoraria from Astellas, AstraZeneca, Novartis, Pharmacyclics, Syndax and Trillium; he is a Chairman of the Scientific Advisory Board of Vincerx Pharmaceuticals and a member of advisory committee of Newave; and is a current equity holder of Vincerx Pharmaceuticals. A-KE is the spouse of Christopher J. Walker who is currently employed by Karyopharm Therapeutics. The remaining authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mrózek, K., Kohlschmidt, J., Blachly, J.S. et al. Outcome prediction by the 2022 European LeukemiaNet genetic-risk classification for adults with acute myeloid leukemia: an Alliance study. Leukemia 37, 788–798 (2023). https://doi.org/10.1038/s41375-023-01846-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01846-8
This article is cited by
-
Outcomes of acute myeloid leukemia patients undergoing allogeneic hematopoietic stem cell transplantation: validation, comparison and improvement of 2022 ELN genetic risk system
Experimental Hematology & Oncology (2024)
-
Stellae-123 gene expression signature improved risk stratification in Taiwanese acute myeloid leukemia patients
Scientific Reports (2024)
-
Validation of the 2022 European LeukemiaNet risk stratification for acute myeloid leukemia
Scientific Reports (2024)
-
A new genomic framework to categorize pediatric acute myeloid leukemia
Nature Genetics (2024)
-
Sex-associated differences in frequencies and prognostic impact of recurrent genetic alterations in adult acute myeloid leukemia (Alliance, AMLCG)
Leukemia (2024)