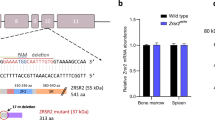
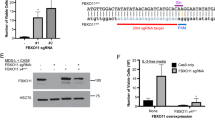
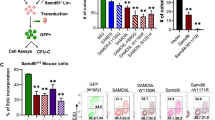
Abstract
Recurrent mutations in RNA splicing proteins and epigenetic regulators contribute to the development of myelodysplastic syndrome (MDS) and related myeloid neoplasms. In chronic myelomonocytic leukemia (CMML), SRSF2 mutations occur in ~50% of patients and TET2 mutations in ~60%. Clonal analysis indicates that either mutation can arise as the founder lesion. Based on human cancer genetics we crossed an inducible Srsf2P95H/+ mutant model with Tet2fl/fl mice to mutate both concomitantly in hematopoietic stem cells. At 20–24 weeks post mutation induction, we observed subtle differences in the Srsf2/Tet2 mutants compared to either single mutant. Under conditions of native hematopoiesis with aging, we see a distinct myeloid bias and monocytosis in the Srsf2/Tet2 mutants. A subset of the compound Srsf2/Tet2 mutants display an increased granulocytic and distinctive monocytic proliferation (myelomonocytic hyperplasia), with increased immature promonocytes and monoblasts and binucleate promonocytes. Exome analysis of progressed disease demonstrated mutations in genes and pathways similar to those reported in human CMML. Upon transplantation, recipients developed leukocytosis, monocytosis, and splenomegaly. We reproduce Srsf2/Tet2 co-operativity in vivo, yielding a disease with core characteristics of CMML, unlike single Srsf2 or Tet2 mutation. This model represents a significant step toward building high fidelity and genetically tractable models of CMML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw data are available in the Sequence Read Archive (SRA) submission: PRJNA850841.
References
Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49:204–12.
Nagata Y, Maciejewski JP. The functional mechanisms of mutations in myelodysplastic syndrome. Leukemia. 2019;33:2779–94.
Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, et al. Combined cohesin–RUNX1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. 2020;10:836–53.
Palomo L, Meggendorfer M, Hutter S, Twardziok S, Adema V, Fuhrmann I, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136:1851–62.
Coltro G, Mangaonkar AA, Lasho TL, Finke CM, Pophali P, Carr R, et al. Clinical, molecular, and prognostic correlates of number, type, and functional localization of TET2 mutations in chronic myelomonocytic leukemia (CMML)-a study of 1084 patients. Leukemia. 2020;34:1407–21.
Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–98.
Meggendorfer M, Haferlach T, Alpermann T, Jeromin S, Haferlach C, Kern W, et al. Specific molecular mutation patterns delineate chronic neutrophilic leukemia, atypical chronic myeloid leukemia, and chronic myelomonocytic leukemia. Haematologica. 2014;99:e244–6.
Orazi A, Germing U. The myelodysplastic/myeloproliferative neoplasms: myeloproliferative diseases with dysplastic features. Leukemia. 2008;22:1308–19.
Pleyer L, Leisch M, Kourakli A, Padron E, Maciejewski JP, Xicoy Cirici B, et al. Outcomes of patients with chronic myelomonocytic leukaemia treated with non-curative therapies: a retrospective cohort study. Lancet. Haematol. 2021;8:e135–48.
Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Ossa JEA, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 2022;1. https://doi.org/10.1056/EVIDoa2200008.
Patnaik MM. How I diagnose and treat chronic myelomonocytic leukemia. Haematologica. 2022;107:1503–17.
Solary E, Itzykson R. How I treat chronic myelomonocytic leukemia. Blood. 2017;130:126–36.
Yoshimi A, Balasis ME, Vedder A, Feldman K, Ma Y, Zhang H, et al. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood. 2017;130:397–407.
Hunter AM, Newman H, Dezern AE, Steensma DP, Niyongere S, Roboz GJ, et al. Integrated human and murine clinical study establishes clinical efficacy of ruxolitinib in chronic myelomonocytic leukemia. Clin Cancer Res. 2021;27:6095–105.
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–59.
Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–30.
Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–3.
Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24.
Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38.
Smeets MF, Tan SY, Xu JJ, Anande G, Unnikrishnan A, Chalk AM, et al. Srsf2 P95H initiates myeloid bias and myelodysplastic/myeloproliferative syndrome from hemopoietic stem cells. Blood. 2018;132:608–21.
Carr RM, Vorobyev D, Lasho T, Marks DL, Tolosa EJ, Vedder A, et al. RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis. Nat Commun. 2021;12:2901.
You X, Liu F, Binder M, Vedder A, Lasho T, Wen Z, et al. Asxl1 loss cooperates with oncogenic Nras in mice to reprogram the immune microenvironment and drive leukemic transformation. Blood. 2022;139:1066–79.
Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18:4872–82.
Gothert JR, Gustin SE, Hall MA, Green AR, Gottgens B, Izon DJ, et al. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 2005;105:2724–32.
Singbrant S, Russell MR, Jovic T, Liddicoat B, Izon DJ, Purton LE, et al. Erythropoietin couples erythropoiesis, B-lymphopoiesis, and bone homeostasis within the bone marrow microenvironment. Blood 2011;117:5631–42.
Smeets MF, DeLuca E, Wall M, Quach JM, Chalk AM, Deans AJ, et al. The Rothmund-Thomson syndrome helicase RECQL4 is essential for hematopoiesis. The. J Clin Investig. 2014;124:3551–65.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Ramsdell JW, Swart JA, Jackson JE, Renvall M. The yield of a home visit in the assessment of geriatric patients. J Am Geriatr Soc. 1989;37:17–24.
Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Kallberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–4.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35.
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92.
Wang HY, Xu X, Ding JH, Bermingham JR Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol Cell. 2001;7:331–42.
Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2018 update on diagnosis, risk stratification and management. Am J Hematol. 2018;93:824–40.
Patel BJ, Przychodzen B, Thota S, Radivoyevitch T, Visconte V, Kuzmanovic T, et al. Genomic determinants of chronic myelomonocytic leukemia. Leukemia 2017;31:2815–23.
An J, González-Avalos E, Chawla A, Jeong M, López-Moyado IF, Li W, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun. 2015;6:10071.
Pan F, Wingo TS, Zhao Z, Gao R, Makishima H, Qu G, et al. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat Commun. 2017;8:15102.
Li Z, Cai X, Cai C-L, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–18.
Nagata Y, Makishima H, Kerr CM, Przychodzen BP, Aly M, Goyal A, et al. Invariant patterns of clonal succession determine specific clinical features of myelodysplastic syndromes. Nat Commun. 2019;10:5386.
Stengel A, Baer C, Walter W, Meggendorfer M, Kern W, Haferlach T, et al. Mutational patterns and their correlation to CHIP-related mutations and age in hematological malignancies. Blood Adv. 2021;5:4426–34.
Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet. 2020;52:378–87.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301.
Acknowledgements
The authors would like to thank L. Purton for discussion and comments; St. Vincent’s Hospital BioResources Centre for care of experimental animals; St. Vincent’s Institute Flow Cytometry Core Facility. This work was supported by the Cancer Council of Victoria (CRW, AMC, MW, and WYL; APP1126010), Victorian Cancer Agency Research Fellowship (CRW; MCRF15015); and in part by the Victorian State Government Operational Infrastructure Support (OIS) to St Vincent’s Institute.
Author information
Authors and Affiliations
Contributions
JJX, MFS, and CRW conceptualized the study; JJX, WYL, MFS, and CRW designed the experiments; JJX, AMC, MW, MFS, and CRW performed the experiments.; JJX and AMC produced the figures; JJX wrote the original manuscript; JJX, AMC, MW, WYL, MFS, and CRW reviewed and edited the manuscript; AMC, MW, WYL, and CRW were responsible for funding acquisition; MFS and CRW provided supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, J.J., Chalk, A.M., Wall, M. et al. Srsf2P95H/+ co-operates with loss of TET2 to promote myeloid bias and initiate a chronic myelomonocytic leukemia-like disease in mice. Leukemia 36, 2883–2893 (2022). https://doi.org/10.1038/s41375-022-01727-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01727-6