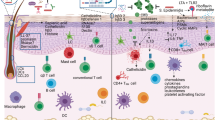
Abstract
The composition of the gut microbiome influences the clinical course after allogeneic hematopoietic stem cell transplantation (HSCT), but little is known about the relevance of skin microorganisms. In a single-center, observational study, we recruited a cohort of 50 patients before undergoing conditioning treatment and took both stool and skin samples up to one year after HSCT. We could confirm intestinal dysbiosis following HSCT and report that the skin microbiome is likewise perturbed in HSCT-recipients. Overall bacterial colonization of the skin was decreased after conditioning. Particularly patients that developed acute skin graft-versus-host disease (aGVHD) presented with an overabundance of Staphylococcus spp. In addition, a loss in alpha diversity was indicative of aGVHD development already before disease onset and correlated with disease severity. Further, co-localization of CD45+ leukocytes and staphylococci was observed in the skin of aGVHD patients even before disease development and paralleled with upregulated genes required for antigen-presentation in mononuclear phagocytes. Overall, our data reveal disturbances of the skin microbiome as well as cutaneous immune response in HSCT recipients with changes associated with cutaneous aGVHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
16S rRNA gene amplicon and whole-genome sequencing data are available under the BioProject accession number PRJNA804670. Bulk RNA sequencing data are deposited in NCBI’s Gene Expression Omnibus database (GSE198625).
Code availability
The code for the co-localization score is available via https://github.com/ramvinay/HostMicrobiome.
Change history
14 February 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41375-023-01839-7
References
Gallo RL. Human Skin Is the Largest Epithelial Surface for Interaction with Microbes. The. J investigative Dermatol. 2017;137:1213–4.
Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–53.
Kabashima K, Honda T, Ginhoux F, Egawa G. The immunological anatomy of the skin. Nat Rev Immunol. 2019;19:19–30.
Brown VI. HSCT-Associated Complications of the Skin, Hair, and Nails. In: Brown VI, editor. Hematopoietic Stem Cell Transplantation for the Pediatric Hematologist/Oncologist. Cham: Springer International Publishing; 2018. p. 363-8.
Zeiser R, Blazar BR. Acute Graft-versus-Host Disease - Biologic Process, Prevention, and Therapy. N. Engl J Med. 2017;377:2167–79.
Strobl J, Pandey RV, Krausgruber T, Bayer N, Kleissl L, Reininger B, et al. Long-term skin-resident memory T cells proliferate in situ and are involved in human graft-versus-host disease. Sci Transl Med. 2020;12:eabb7028.
Strong Rodrigues K, Oliveira-Ribeiro C, de Abreu Fiuza Gomes S, Knobler R. Cutaneous Graft-Versus-Host Disease: Diagnosis and Treatment. Am J Clin Dermatol. 2018;19:33–50.
Mathewson N, Reddy P. The Microbiome and Graft Versus Host Disease. Curr Stem Cell Rep. 2015;1:39–47.
Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–82.
Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl J Med. 2020;382:822–34.
Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–55.
Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. 2021;27:700–9.
Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18:295–304.
Filipovich AH, Weisdorf D, Pavletic S, Socie G, Wingard JR, Lee SJ, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transpl. 2005;11:945–56.
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. Isme j. 2011;5:1571–9.
Pjevac P, Hausmann B, Schwarz J, Kohl G, Herbold CW, Loy A, et al. An Economical and Flexible Dual Barcoding, Two-Step PCR Approach for Highly Multiplexed Amplicon Sequencing. Front Microbiol. 2021;12:669776.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Res. 2016;5:1492.
Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Kasper S, Andersen RHK, Søren M. Karst, Mads Albertsen. mpvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv. 2019. https://github.com/MadsAlbertsen/ampvis2.
Jari Oksanen FGB, Michael F, Roeland K, Pierre L, Dan McGlinn, Peter R, et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. http://CRAN.Rproject.org/package=vegan.
Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Micro. 2008;6:339–48.
Trebesius K, Leitritz L, Adler K, Schubert S, Autenrieth IB, Heesemann J. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med Microbiol Immunol. 2000;188:169–75.
Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–13.
Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–81.
Gómez-Sánchez D, Schlötterer C. ReadTools: A universal toolkit for handling sequence data from different sequencing platforms. Mol Ecol Resour. 2018;18:676–80.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Brüggen M-C, Klein I, Greinix H, Bauer W, Kuzmina Z, Rabitsch W, et al. Diverse T-cell responses characterize the different manifestations of cutaneous graft-versus-host disease. Blood. 2014;123:290–9.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–2.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinforma. 2011;12:124.
Casson CN, Yu J, Reyes VM, Taschuk FO, Yadav A, Copenhaver AM, et al. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc Natl Acad Sci. 2015;112:6688.
Alloatti A, Kotsias F, Pauwels A-M, Carpier J-M, Jouve M, Timmerman E, et al. Toll-like Receptor 4 Engagement on Dendritic Cells Restrains Phago-Lysosome Fusion and Promotes Cross-Presentation of Antigens. Immunity. 2015;43:1087–100.
Buschow SI, Lasonder E, Szklarczyk R, Oud MM, de Vries IJM, Figdor CG. Unraveling the human dendritic cell phagosome proteome by organellar enrichment ranking. J Proteom. 2012;75:1547–62.
Li S, Paulsson KM, Chen S, Sjögren HO, Wang P. Tapasin is required for efficient peptide binding to transporter associated with antigen processing. J Biol Chem. 2000;275:1581–6.
Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, et al. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–71.
Simmons DP, Wearsch PA, Canaday DH, Meyerson HJ, Liu YC, Wang Y, et al. Type I IFN Drives a Distinctive Dendritic Cell Maturation Phenotype That Allows Continued Class II MHC Synthesis and Antigen Processing. J Immunol. 2012;188:3116–26.
Gérard A, Patino-Lopez G, Beemiller P, Nambiar R, Ben-Aissa K, Liu Y, et al. Detection of rare antigen-presenting cells through T cell-intrinsic meandering motility, mediated by Myo1g. Cell. 2014;158:492–505.
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484–506.
Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K, et al. Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs Host Disease After Hematopoietic Cell Transplantation. Clin Infect Dis. 2017;65:1984–91.
Spindelboeck W, Schulz E, Uhl B, Kashofer K, Aigelsreiter A, Zinke-Cerwenka W, et al. Repeated fecal microbiota transplantations attenuate diarrhea and lead to sustained changes in the fecal microbiota in acute, refractory gastrointestinal graft-versus-host-disease. Haematologica. 2017;102:e210–3.
van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci Transl Med. 2020;12:eaaz8926.
Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transpl. 2014;20:640–5.
Biagi E, Zama D, Nastasi C, Consolandi C, Fiori J, Rampelli S, et al. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transpl. 2015;50:992–8.
Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transpl. 2015;21:1373–83.
Gu Y, Sun J, Li K, Wu X, Zhang J. Alteration in the Skin Microbiome in Cutaneous Graft Versus Host Disease. Acta Derm Venereol. 2021;101:adv00374.
Ghimire S, Weber D, Mavin E, Wang Xn, Dickinson AM, Holler E. Pathophysiology of GvHD and Other HSCT-Related Major Complications. Front Immunol. 2017;8:79.
Lei YM, Sepulveda M, Chen L, Wang Y, Pirozzolo I, Theriault B, et al. Skin-restricted commensal colonization accelerates skin graft rejection. JCI Insight. 2019;5:e127569.
Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JLM. Total Body Irradiation and Acute Graft-Versus-Host Disease: The Role of Gastrointestinal Damage and Inflammatory Cytokines. Blood. 1997;90:3204–13.
Nestel FP, Price KS, Seemayer TA, Lapp WS. Macrophage priming and lipopolysaccharide-triggered release of tumor necrosis factor alpha during graft-versus-host disease. J Exp Med. 1992;175:405–13.
Penack O, Smith OM, Cunningham-Bussel A, Liu X, Rao U, Yim N, et al. NOD2 regulates hematopoietic cell function during graft-versus-host disease. J Exp Med. 2009;206:2101–10.
Heimesaat MM, Nogai A, Bereswill S, Plickert R, Fischer A, Loddenkemper C, et al. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut. 2010;59:1079–87.
Williams MR, Cau L, Wang Y, Kaul D, Sanford JA, Zaramela LS, et al. Interplay of Staphylococcal and Host Proteases Promotes Skin Barrier Disruption in Netherton Syndrome. Cell Rep. 2020;30:2923–33.e7.
Acknowledgements
We thank the Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna as well as Biomedical Sequencing Facility (BSF) at CeMM for assistance with next-generation sequencing. This work was supported by funds of the Austrian Science Fund (FWF, P31494), the Oesterreichische Nationalbank (Austrian Central Bank; Anniversary Fund; project number, 17872), and by the Innovation Fund of the Austrian Academy of Sciences (ÖAW; project number, IF_2017_29). NB was supported by the ESCMID Research Grant 2020 of the European Society of Clinical Microbiology and Infectious Diseases. SK received funding from the Austrian Science Fund, FWF, via the Doctoral Program Cell Communication in Health and Disease (W 1205-B09), and the Special Research Programs Chromatin Landscapes (F6104) and Immunothrombosis (F5410).
Author information
Authors and Affiliations
Contributions
Conceptualization, GS; Methodology, NB; Software, RN and MLW; Formal Analysis, NB, BH, RVP, FD, PP, CK and RK; Investigation, NB, LMG, JS, AR, VB, LK, MN and LH; Resources, CS, PW and WR; Data Curation, NB, BH and LU; Writing—Original Draft, NB.; Writing—Review and Editing, GS, SK; DB and AM; Visualization, NB; Supervision, GS, SK; DB and RCE; Funding Acquisition, GS, NB.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The author’s name Bela Hausmann has been corrected.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bayer, N., Hausmann, B., Pandey, R.V. et al. Disturbances in microbial skin recolonization and cutaneous immune response following allogeneic stem cell transfer. Leukemia 36, 2705–2714 (2022). https://doi.org/10.1038/s41375-022-01712-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01712-z