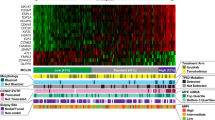
Abstract
Interrogation of cell-free DNA (cfDNA) represents an emerging approach to non-invasively estimate disease burden in multiple myeloma (MM). Here, we examined low-pass whole genome sequencing (LPWGS) of cfDNA for its predictive value in relapsed/ refractory MM (RRMM). We observed that cfDNA positivity, defined as ≥10% tumor fraction by LPWGS, was associated with significantly shorter progression-free survival (PFS) in an exploratory test cohort of 16 patients who were actively treated on diverse regimens. We prospectively determined the predictive value of cfDNA in 86 samples from 45 RRMM patients treated with elotuzumab, pomalidomide, bortezomib, and dexamethasone in a phase II clinical trial (NCT02718833). PFS in patients with tumor-positive and -negative cfDNA after two cycles of treatment was 1.6 and 17.6 months, respectively (HR 7.6, P < 0.0001). Multivariate hazard modelling confirmed cfDNA as independent risk factor (HR 96.6, P = 6.92e-05). While correlating with serum-free light chains and bone marrow, cfDNA additionally discriminated patients with poor PFS among those with the same response by IMWG criteria. In summary, detectability of MM-derived cfDNA, as a measure of substantial tumor burden with therapy, independently predicts poor PFS and may provide refinement for standard-of-care response parameters to identify patients with poor response to treatment earlier than is currently feasible.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–28.
Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–76.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–38.
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72.
Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48.
Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–20.
Kumar SK, Therneau TM, Gertz MA, Lacy MQ, Dispenzieri A, Rajkumar SV, et al. Clinical course of patients with relapsed multiple myeloma. Mayo Clin Proc. 2004;79:867–74.
Landgren O, Rajkumar SV. New Developments in Diagnosis, Prognosis, and Assessment of Response in Multiple Myeloma. Clin Cancer Res. 2016;22:5428–33.
Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268.
Caers J, Paiva B, Zamagni E, Leleu X, Bladé J, Kristinsson SY, et al. Diagnosis, treatment, and response assessment in solitary plasmacytoma: updated recommendations from a European Expert Panel. J Hematol Oncol. 2018;11:10.
Chawla SS, Kumar SK, Dispenzieri A, Greenberg AJ, Larson DR, Kyle RA et al. Clinical Course and Prognosis of Non-Secretory Multiple Myeloma. Eur J Haematol. 2015. https://doi.org/10.1111/ejh.12534.
Dupuis MM, Tuchman SA. Non-secretory multiple myeloma: from biology to clinical management. OncoTargets Ther. 2016;9:7583–90.
Mills JR, Barnidge DR, Dispenzieri A, Murray DL. High sensitivity blood-based M-protein detection in sCR patients with multiple myeloma. Blood Cancer J. 2017;7:e590.
Mithraprabhu S, Chen M, Savvidou I, Reale A, Spencer A Liquid biopsy: an evolving paradigm for the biological characterisation of plasma cell disorders. Leukemia 2021. https://doi.org/10.1038/s41375-021-01339-6.
Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017; 8. https://doi.org/10.1038/s41467-017-00965-y.
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14.
Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Scherer F, Kurtz DM, Newman AM, Stehr H, Craig AFM, Esfahani MS, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8:364ra155.
Corcoran RB, Chabner BA. Cell-free DNA Analysis in Cancer. N. Engl J Med. 2019;380:501–2.
Manier S, Park J, Capelletti M, Bustoros M, Freeman SS, Ha G, et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat Commun. 2018;9:1691.
Guo G, Raje NS, Seifer C, Kloeber J, Isenhart R, Ha G, et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia. 2018;32:1838–41.
Gerber B, Manzoni M, Spina V, Bruscaggin A, Lionetti M, Fabris S, et al. Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica. 2018;103:e245–8.
Mithraprabhu S, Khong T, Ramachandran M, Chow A, Klarica D, Mai L, et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia. 2017;31:1695–705.
Mithraprabhu S, Morley R, Khong T, Kalff A, Bergin K, Hocking J, et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia. 2019;33:2022–33.
Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017;8:15086.
Rustad EH, Coward E, Skytøen ER, Misund K, Holien T, Standal T, et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica. 2017;102:1266–72.
Li Q, Huang HJ, Ma J, Wang Y, Cao Z, Karlin-Neumann G, et al. RAS/RAF mutations in tumor samples and cell-free DNA from plasma and bone marrow aspirates in multiple myeloma patients. J Cancer. 2020;11:3543–50.
Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica. 2017;102:1105–11.
Biancon G, Gimondi S, Vendramin A, Carniti C, Corradini P. Noninvasive molecular monitoring in multiple myeloma patients using cell-free tumor DNA: A pilot study. J Mol Diagn. 2018;20:859–70.
Mazzotti C, Buisson L, Maheo S, Perrot A, Chretien M-L, Leleu X, et al. Myeloma MRD by deep sequencing from circulating tumor DNA does not correlate with results obtained in the bone marrow. Blood Adv. 2018;2:2811–3.
Deshpande S, Tytarenko RG, Wang Y, Boyle EM, Ashby C, Schinke CD, et al. Monitoring treatment response and disease progression in myeloma with circulating cell-free DNA. Eur J Haematol. 2021;106:230–40.
Mithraprabhu S, Sirdesai S, Chen M, Khong T, Spencer A. Circulating tumour DNA analysis for tumour genome characterisation and monitoring disease burden in extramedullary multiple myeloma. Int J Mol Sci. 2018; 19. https://doi.org/10.3390/ijms19071858.
Manzoni M, Pompa A, Fabris S, Pelizzoni F, Ciceri G, Seia M, et al. Limits and applications of genomic analysis of circulating tumor DNA as a liquid biopsy in asymptomatic forms of multiple myeloma. HemaSphere. 2020;4:e402.
Yee AJ, Laubach JP, Campagnaro EL, Lipe BC, Nadeem O, Friedman RS, et al. A phase II study of elotuzumab in combination with pomalidomide, bortezomib, and dexamethasone in relapsed and refractory multiple myeloma. Blood. 2019;134:3169.
Ihorst G, Waldschmidt J, Schumacher M, Wäsch R, Engelhardt M. Analysis of survival by tumor response: have we learnt any better? Ann Hematol. 2015;94:1615–6.
Chng WJ, Dispenzieri A, Chim C-S, Fonseca R, Goldschmidt H, Lentzsch S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–77.
Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau J-L, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585–90.
Kortuem KM, Braggio E, Bruins L, Barrio S, Shi CS, Zhu YX, et al. Panel sequencing for clinically oriented variant screening and copy number detection in 142 untreated multiple myeloma patients. Blood Cancer J. 2016;6:e397.
Frede J, Anand P, Sotudeh N, Pinto RA, Nair MS, Stuart H et al. Dynamic transcriptional reprogramming leads to immunotherapeutic vulnerabilities in myeloma. Nat Cell Biol. 2021. https://doi.org/10.1038/s41556-021-00766-y.
Waldschmidt JM, Kloeber JA, Anand P, Frede J, Kokkalis A, Dimitrova V et al. Single-cell profiling reveals metabolic reprogramming as a resistance mechanism in BRAF-mutated multiple myeloma. Clin Cancer Res. 2021. https://doi.org/10.1158/1078-0432.CCR-21-2040.
Vrabel D, Sedlarikova L, Besse L, Rihova L, Bezdekova R, Almasi M, et al. Dynamics of tumor-specific cfDNA in response to therapy in multiple myeloma patients. Eur J Haematol. 2020;104:190–7.
Engelhardt M, Ihorst G, Duque-Afonso J, Wedding U, Spät-Schwalbe E, Goede V, et al. Structured assessment of frailty in multiple myeloma as a paradigm of individualized treatment algorithms in cancer patients at advanced age. Haematologica. 2020;105:1183–8.
Alidousty C, Brandes D, Heydt C, Wagener S, Wittersheim M, Schäfer SC, et al. Comparison of blood collection tubes from three different manufacturers for the collection of cell-free DNA for liquid biopsy mutation testing. J Mol Diagn. 2017;19:801–4.
Mithraprabhu S, Spencer A. Analysis of circulating tumor DNA. Methods Mol Biol. 2018;1792:129–45.
Bianchi G, Kyle RA, Larson DR, Witzig TE, Kumar S, Dispenzieri A, et al. High levels of peripheral blood circulating plasma cells as a specific risk factor for progression of smoldering multiple myeloma. Leukemia. 2013;27:680–5.
Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8:363ra147.
Mishima Y, Paiva B, Shi J, Park J, Manier S, Takagi S, et al. The mutational landscape of circulating tumor cells in multiple myeloma. Cell Rep. 2017;19:218–24.
Kumar S, Rajkumar SV, Kyle RA, Lacy MQ, Dispenzieri A, Fonseca R, et al. Prognostic value of circulating plasma cells in monoclonal gammopathy of undetermined significance. J Clin Oncol. 2005;23:5668–74.
Gonsalves WI, Rajkumar SV, Dispenzieri A, Dingli D, Timm MM, Morice WG, et al. Quantification of circulating clonal plasma cells via multiparametric flow cytometry identifies patients with smoldering multiple myeloma at high risk of progression. Leukemia. 2017;31:130–5.
Gonsalves WI, Jevremovic D, Nandakumar B, Dispenzieri A, Buadi FK, Dingli D, et al. Enhancing the R-ISS classification of newly diagnosed multiple myeloma by quantifying circulating clonal plasma cells. Am J Hematol. 2020;95:310–5.
Paiva B, Paino T, Sayagues J-M, Garayoa M, San-Segundo L, Martín M, et al. Detailed characterization of multiple myeloma circulating tumor cells shows unique phenotypic, cytogenetic, functional, and circadian distribution profile. Blood. 2013;122:3591–8.
Sanoja-Flores L, Flores-Montero J, Puig N, Contreras-Sanfeliciano T, Pontes R, Corral-Mateos A, et al. Blood monitoring of circulating tumor plasma cells by next generation flow in multiple myeloma after therapy. Blood. 2019;134:2218–22.
Garcés J-J, Bretones G, Burgos L, Valdes-Mas R, Puig N, Cedena M-T, et al. Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia. 2020;34:3007–18.
Kubiczkova L, Kryukov F, Slaby O, Dementyeva E, Jarkovsky J, Nekvindova J, et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99:511–8.
Wang W, Corrigan-Cummins M, Barber EA, Saleh LM, Zingone A, Ghafoor A, et al. Aberrant levels of miRNAs in bone marrow microenvironment and peripheral blood of myeloma patients and disease progression. J Mol Diagn. 2015;17:669–78.
Navarro A, Díaz T, Tovar N, Pedrosa F, Tejero R, Cibeira MT, et al. A serum microRNA signature associated with complete remission and progression after autologous stem-cell transplantation in patients with multiple myeloma. Oncotarget. 2015;6:1874–83.
Lionetti M, Musto P, Di Martino MT, Fabris S, Agnelli L, Todoerti K, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res. 2013;19:3130–42.
Acknowledgements
JMW is supported by a postdoctoral fellowship of Deutsche Forschungsgemeinschaft (German Research Foundation, 391926441) and the International Myeloma Society Young Investigator Award (IMW, Boston, 2019). JGL is supported by the NCI (K08CA191026), the V Foundation for Cancer Research, and the Anna Fuller Fund. BK is supported by the NCI (K08CA191091).
Author information
Authors and Affiliations
Contributions
JMW, TV, SP, BK, and JGL designed and performed experiments and analyzed the data. AJY, NSR, JGL, MM, LB, GB, SS, RF, BL, EC, ED, JPL, NCM, PGR, and KCA designed the study and provided clinical data analysis. JMW, TV, RPR, PA, CZZ, BK and JGL conceived and implemented computational methods for data analysis. JF, SP, MSN, AK, GG, and JK provided analytical support. JMW, AJY, TV, NSR, BK, and JGL wrote the paper. JGL, BK, and NSR designed the experimental strategy and supervised the analysis. All authors discussed the results and implications and reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
No disclosures are related to this publication. JMW: Advisory boards of Janssen and Sanofi. AJY: Advisory boards of Adaptive, Amgen, Bristol-Myers Squibb, GSK, Janssen, Karyopharm, Oncopeptides, Regeneron, Sanofi and Takeda. Clinical trial support from Adaptive, Amgen, BMS, Bristol-Myers Squibb, Janssen and Takeda. BL: Research funding from Amgen and Cellectar. Advisory boards of Bristol-Myers Squibb, Janssen, and GlaxoSmithKline. C-ZZ: Co-founder, advisor, and equity holder of Pillar BioSciences. PGR: Advisory boards of AbbVie, AstraZeneca, GSK, Bristol-Myers Squibb, Oncopeptides, Celgene, Takeda, and Karyopharm. Advisory boards of Oncopeptides, Janssen, Sanofi, and Secura Bio. KCA: Advisory boards of Janssen, Amgen, Pfizer, AstraZeneca, Precision Biosciences, Bristol-Myers Squibb, Mana, Starton, Window during the conduct of the study; other support from C4 Therapeutics, Oncopep, Raqia; and other support from NextRNA outside the submitted work. NSR: Advisory boards of Amgen, Bristol-Myers Squibb, Janssen, Sanofi, Takeda, AstraZeneca, GSK, Pfizer, Caribou, Immuneel and C4 Therapeutics. Research funding from BluebirdBio. JGL: Consultant for T2 Biosystems outside the submitted work. Research funding from Bristol-Myers Squibb, Celgene. All other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Introduction
Supplementary information
Rights and permissions
About this article
Cite this article
Waldschmidt, J.M., Yee, A.J., Vijaykumar, T. et al. Cell-free DNA for the detection of emerging treatment failure in relapsed/ refractory multiple myeloma. Leukemia 36, 1078–1087 (2022). https://doi.org/10.1038/s41375-021-01492-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01492-y
This article is cited by
-
Liquid biopsy by analysis of circulating myeloma cells and cell-free nucleic acids: a novel noninvasive approach of disease evaluation in multiple myeloma
Biomarker Research (2023)
-
Cell-free DNA chromosome copy number variations predict outcomes in plasma cell myeloma
Blood Cancer Journal (2023)
-
An Investigation into Cell-Free DNA in Different Common Cancers
Molecular Biotechnology (2023)
-
Response-Adapted Therapy for Newly Diagnosed Multiple Myeloma
Current Hematologic Malignancy Reports (2023)