Abstract
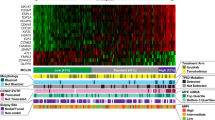
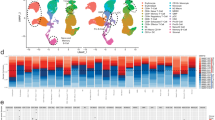
Mantle cell lymphoma (MCL) is characterized by marked differences in outcome, emphasizing the need for strong prognostic biomarkers. Here, we explore expression patterns and prognostic relevance of circular RNAs (circRNAs), a group of endogenous non-coding RNA molecules, in MCL. We profiled the circRNA expression landscape using RNA-sequencing and explored the prognostic potential of 40 abundant circRNAs in samples from the Nordic MCL2 and MCL3 clinical trials, using NanoString nCounter Technology. We report a circRNA-based signature (circSCORE) developed in the training cohort MCL2 that is highly predictive of time to progression (TTP) and lymphoma-specific survival (LSS). The dismal outcome observed in the large proportion of patients assigned to the circSCORE high-risk group was confirmed in the independent validation cohort MCL3, both in terms of TTP (HR 3.0; P = 0.0004) and LSS (HR 3.6; P = 0.001). In Cox multiple regression analysis incorporating MIPI, Ki67 index, blastoid morphology and presence of TP53 mutations, circSCORE retained prognostic significance for TTP (HR 3.2; P = 0.01) and LSS (HR 4.6; P = 0.01). In conclusion, circRNAs are promising prognostic biomarkers in MCL and circSCORE improves identification of high-risk disease among younger patients treated with cytarabine-containing chemoimmunotherapy and autologous stem cell transplant.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Codes used for data analyses can be obtained from the corresponding author upon request.
Change history
18 February 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41375-022-01526-z
References
Geisler CH, Kolstad A, Laurell A, Jerkeman M, Räty R, Andersen NS, et al. Nordic MCL2 trial update: six-year follow-up after intensive immunochemotherapy for untreated mantle cell lymphoma followed by BEAM or BEAC+ autologous stem-cell support: still very long survival but late relapses do occur. Br J Haematol. 2012;158:355–62.
Hermine O, Hoster E, Walewski J, Bosly A, Stilgenbauer S, Thieblemont C, et al. Addition of high-dose cytarabine to immunochemotherapy before autologous stem-cell transplantation in patients aged 65 years or younger with mantle cell lymphoma (MCL younger): a randomised, open-label, phase 3 trial of the European Mantle Cell Lymphoma Network. Lancet. 2016;388:565–75.
Chihara D, Cheah CY, Westin JR, Fayad LE, Rodriguez MA, Hagemeister FB, et al. Rituximab plus hyper-CVAD alternating with MTX/Ara-C in patients with newly diagnosed mantle cell lymphoma: 15-year follow-up of a phase II study from the MD Anderson Cancer Center. Br J Haematol. 2016;172:80–8.
Geisler CH, Kolstad A, Laurell A, Andersen NS, Pedersen LB, Jerkeman M, et al. Long-term progression-free survival of mantle cell lymphoma after intensive front-line immunochemotherapy with in vivo-purged stem cell rescue: a nonrandomized phase 2 multicenter study by the Nordic Lymphoma Group. Blood. 2008;112:2687–93.
Dreyling M, Geisler C, Hermine O, Kluin-Nelemans HC, Le Gouill S, Rule S, et al. Newly diagnosed and relapsed mantle cell lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;1:iv62–71.
Eskelund CW, Kolstad A, Jerkeman M, Räty R, Laurell A, Eloranta S, et al. 15-year follow-up of the Second Nordic Mantle Cell Lymphoma trial (MCL2): prolonged remissions without survival plateau. Br J Haematol. 2016;175:410–8.
Hoster E, Dreyling M, Klapper W, Gisselbrecht C, Van Hoof A, Kluin-Nelemans HC, et al. A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558–65.
Tiemann M, Schrader C, Klapper W, Dreyling MH, Campo E, Norton A, et al. Histopathology, cell proliferation indices and clinical outcome in 304 patients with mantle cell lymphoma (MCL): a clinicopathological study from the European MCL Network. Br J Haematol. 2005;131:29–38.
Hoster E, Rosenwald A, Berger F, Bernd H-W, Hartmann S, Loddenkemper C, et al. Prognostic value of Ki-67 index, cytology, and growth pattern in mantle-cell lymphoma: results from randomized trials of the European Mantle Cell Lymphoma Network. J Clin Oncol. 2016;34:1386–94.
Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–97.
Scott DW, Abrisqueta P, Wright GW, Slack GW, Mottok A, Villa D, et al. New molecular assay for the proliferation signature in mantle cell lymphoma applicable to formalin-fixed paraffin-embedded biopsies. J Clin Oncol. 2017;35:1668–77.
Husby S, Ralfkiaer U, Garde C, Zandi R, Ek S, Kolstad A, et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood. 2015;125:2669–77.
Beà S, Amador V. Role of SOX11 and genetic events cooperating with cyclin D1 in mantle cell lymphoma. Curr Oncol Rep. 2017;19:43.
Pararajalingam P, Coyle KM, Arthur SE, Thomas N, Alcaide M, Meissner B, et al. Coding and noncoding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood. 2020;136:572–84.
Nadeu F, Martin-Garcia D, Clot G, Díaz-Navarro A, Duran-Ferrer M, Navarro A, et al. Genomic and epigenomic insights into the origin, pathogenesis, and clinical behavior of mantle cell lymphoma subtypes. Blood. 2020;136:1419–32.
Eskelund CW, Dahl C, Hansen JW, Westman M, Kolstad A, Pedersen LB, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130:1903–10.
Ferrero S, Rossi D, Rinaldi A, Bruscaggin A, Spina V, Eskelund CW, et al. KMT2D mutations and TP53 disruptions are poor prognostic biomarkers in mantle cell lymphoma receiving high-dose therapy: a FIL study. Haematologica. 2020;105:1604–12.
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733.
Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95:1179–89.
Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J-L, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–24.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2017;37:555–65.
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–92.
Chen DF, Zhang LJ, Tan K, Jing Q. Application of droplet digital PCR in quantitative detection of the cell-free circulating circRNAs. Biotechnol Biotechnol Equip. 2018;32:116–23.
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25.
Dahl M, Daugaard I, Andersen M, Hansen TB, Grønbæk K, Kjems J, et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab Invest. 2018;98:1657–69.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. CircRNA biogenesis competes with Pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–9.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Kolstad A, Laurell A, Jerkeman M, Grønbæk K, Elonen E, Räty R, et al. Nordic MCL-3 study: BEAM/C conditioning intensified with 90Y-ibritumomab-tiuxetan in responding non-CR patients followed by autologous transplant in mantle cell lymphoma. Blood. 2014;123:2953–9.
Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245.
Ogłuszka M, Orzechowska M, Jędroszka D, Witas P, Bednarek AK. Evaluate cutpoints: adaptable continuous data distribution system for determining survival in Kaplan-Meier estimator. Comput Methods Programs Biomed. 2019;177:133–9.
Glazar P, Papavasileiou P, Rajewsky N, Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70.
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47.
Hansen TB, Venø MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2015;44:e58.
Kristensen LS, Ebbesen KK, Sokol M, Jakobsen T, Korsgaard U, Eriksen AC, et al. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat Commun. 2020;11:4551.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.
Wlodarska I, Veyt E, De Paepe P, Vandenberghe P, Nooijen P, Theate I, et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia. 2005;19:1299–305.
Licht JD. SETD2: a complex role in blood malignancy. Blood. 2017;130:2576–8.
Agarwal R, Chan Y-C, Tam CS, Hunter T, Vassiliadis D, Teh CE, et al. Dynamic molecular monitoring reveals that SWI–SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med. 2019;25:119–29.
Moldovan L-I, Hansen TB, Venø MT, Okholm TLH, Andersen TL, Hager H, et al. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med Genomics. 2019;12:174.
Das Mahapatra K, Pasquali L, Søndergaard JN, Lapins J, Nemeth IB, Baltás E, et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci Rep. 2020;10:3637.
Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057.
Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234:10646–54.
Zhu L, Liu Y, Yang Y, Mao X-M, Yin Z-D. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23:2817–26.
Liu Z, Pan H-M, Xin L, Zhang Y, Zhang W-M, Cao P, et al. Circ-ZNF609 promotes carcinogenesis of gastric cancer cells by inhibiting miRNA-145-5p expression. Eur Rev Med Pharmacol Sci. 2019;23:9411–7.
Liao X, Zhan W, Tian B, Luo Y, Gu F, Li R. Circular RNA ZNF609 promoted hepatocellular carcinoma progression by upregulating PAP2C expression via sponging miR-342-3p. OncoTargets Ther. 2020;13:7773–83.
Rossi F, Legnini I, Megiorni F, Colantoni A, Santini T, Morlando M, et al. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38:3843–54.
Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol. 2020;10:982.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Klapper W, Hoster E, Determann O, Oschlies I, van der Laak J, Berger F, et al. Ki-67 as a prognostic marker in mantle cell lymphoma—consensus guidelines of the pathology panel of the European MCL Network. J Hematop. 2009;2:103–11.
Croci GA, Hoster E, Beà S, Clot G, Enjuanes A, Scott DW, et al. Reproducibility of histologic prognostic parameters for mantle cell lymphoma: cytology, Ki67, p53 and SOX11. Virchows Arch. 2020;477:259–67.
Ramsower CA, Maguire A, Robetorye RS, Feldman AL, Syrbu SI, Rosenthal AC, et al. Clinical laboratory validation of the MCL35 assay for molecular risk stratification of mantle cell lymphoma. J Hematop. 2020;13:231–8.
Holte H, Beiske K, Boyle M, Trøen G, Blaker YN, Myklebust J, et al. The MCL35 gene expression proliferation assay predicts high-risk MCL patients in a Norwegian cohort of younger patients given intensive first line therapy. Br J Haematol. 2018;183:225–34.
Rauert-Wunderlich H, Mottok A, Scott DW, Rimsza LM, Ott G, Klapper W, et al. Validation of the MCL35 gene expression proliferation assay in randomized trials of the European Mantle Cell Lymphoma Network. Br J Haematol. 2019;184:616–24.
Acknowledgements
The authors would like to thank staff members of all contributing departments of the MCL2 and MCL3 clinical trials, and we are grateful to all patients who participated. We thank Morten Venø for his help processing the RNA-seq data and MD Eileen Wedge for proofreading the manuscript. This work was supported by grants from Rigshospitalets Research foundation, The Novo Nordisk Foundation (NNF17OC0029596), The Lundbeck Foundation (R307-2018-3433), “Mindegaard Donationen, Rigshospitalet” and “Sejer Persson & Lis Klüver Persson’s fund”. K.G is funded by a center grant from The Danish Cancer Society (Danish Research Center for Precision Medicine in Blood Cancer; grant 223-A13071-18-S68), from the Novo Nordisk Foundation (Novo Nordisk Foundation Center for Stem Cell Biology, DanStem; grant NNF17CC0027852) and from Greater Copenhagen Health Science Partners (Clinical Academic Group in Blood Cancers).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dahl, M., Husby, S., Eskelund, C.W. et al. Expression patterns and prognostic potential of circular RNAs in mantle cell lymphoma: a study of younger patients from the MCL2 and MCL3 clinical trials. Leukemia 36, 177–188 (2022). https://doi.org/10.1038/s41375-021-01311-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01311-4
This article is cited by
-
Circular ZDHHC11 supports Burkitt lymphoma growth independent of its miR-150 binding capacity
Scientific Reports (2024)
-
The role of circular RNA during the urological cancer metastasis: exploring regulatory mechanisms and potential therapeutic targets
Cancer and Metastasis Reviews (2024)
-
Impact of U2AF1 mutations on circular RNA expression in myelodysplastic neoplasms
Leukemia (2023)
-
Going circular: history, present, and future of circRNAs in cancer
Oncogene (2023)
-
Defining the landscape of circular RNAs in neuroblastoma unveils a global suppressive function of MYCN
Nature Communications (2023)