Abstract
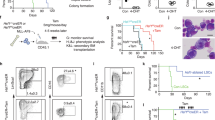
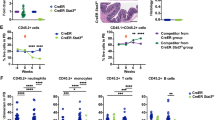
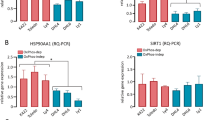
Shwachman–Diamond syndrome (SDS) is a bone marrow failure (BMF) syndrome associated with an increased risk of myelodysplasia and leukemia. The molecular mechanisms of SDS are not fully understood. We report that primitive hematopoietic cells from SDS patients present with a reduced activity of the small RhoGTPase Cdc42 and concomitantly a reduced frequency of HSCs polar for polarity proteins. The level of apolarity of SDS HSCs correlated with the magnitude of HSC depletion in SDS patients. Importantly, exogenously provided Wnt5a or GDF11 that elevates the activity of Cdc42 restored polarity in SDS HSCs and increased the number of HSCs in SDS patient samples in surrogate ex vivo assays. Single cell level RNA-Seq analyses of SDS HSCs and daughter cells demonstrated that SDS HSC treated with GDF11 are transcriptionally more similar to control than to SDS HSCs. Treatment with GDF11 reverted pathways in SDS HSCs associated with rRNA processing and ribosome function, but also viral infection and immune function, p53-dependent DNA damage, spindle checkpoints, and metabolism, further implying a role of these pathways in HSC failure in SDS. Our data suggest that HSC failure in SDS is driven at least in part by low Cdc42 activity in SDS HSCs. Our data thus identify novel rationale approaches to attenuate HSCs failure in SDS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Myers KC, Davies SM, Shimamura A. Clinical and molecular pathophysiology of Shwachman-Diamond syndrome: an update. Hematol Oncol Clin North Am. 2013;27:117–28.
Huang JN, Shimamura A. Clinical spectrum and molecular pathophysiology of Shwachman-Diamond syndrome. Curr Opin Hematol. 2011;18:30–5.
Hashmi SK, Allen C, Klaassen R, Fernandez CV, Yanofsky R, Shereck E, et al. Comparative analysis of Shwachman-Diamond syndrome to other inherited bone marrow failure syndromes and genotype-phenotype correlation. Clin Genet. 2011;79:448–58.
Donadieu J, Fenneteau O, Beaupain B, Beaufils S, Bellanger F, Mahlaoui N, et al. Classification of and risk factors for hematologic complications in a French national cohort of 102 patients with Shwachman-Diamond syndrome. Haematologica. 2012;97:1312–9.
Dror Y, Freedman MH. Shwachman-Diamond syndrome: an inherited preleukemic bone marrow failure disorder with aberrant hematopoietic progenitors and faulty marrow microenvironment. Blood. 1999;94:3048–54.
Andre V, Longoni D, Bresolin S, Cappuzzello C, Dander E, Galbiati M, et al. Mesenchymal stem cells from Shwachman-Diamond syndrome patients display normal functions and do not contribute to hematological defects. Blood Cancer J. 2012;2:e94.
Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101.
Dhanraj S, Matveev A, Li H, Lauhasurayotin S, Jardine L, Cada M, et al. Biallelic mutations in DNAJC21 cause Shwachman-Diamond syndrome. Blood. 2017;129:1557–62.
Stepensky P, Chacon-Flores M, Kim KH, Abuzaitoun O, Bautista-Santos A, Simanovsky N, et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in aShwachman-Diamond like syndrome. J Med Genet. 2017;54:558–66.
Carapito R, Konantz M, Paillard C, Miao Z, Pichot A, Leduc MS, et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J Clin Invest. 2017;127:4090–103.
Rujkijyanont P, Adams SL, Beyene J, Dror Y. Bone marrow cells from patients with Shwachman-Diamond syndrome abnormally express genes involved in ribosome biogenesis and RNA processing. Br J Haematol. 2009;145:806–15.
Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–12.
Burwick N, Coats SA, Nakamura T, Shimamura A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood. 2012;120:5143–52.
Zhang S, Shi M, Hui CC, Rommens JM. Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol. 2006;26:6656–63.
Rawls AS, Gregory AD, Woloszynek JR, Liu F, Link DC. Lentiviral-mediated RNAi inhibition of Sbds in murine hematopoietic progenitors impairs their hematopoietic potential. Blood. 2007;110:2414–22.
Zambetti NA, Bindels EM, Van Strien PM, Valkhof MG, Adisty MN, Hoogenboezem RM, et al. Deficiency of the ribosome biogenesis gene Sbds in hematopoietic stem and progenitor cells causes neutropenia in mice by attenuating lineage progression in myelocytes. Haematologica. 2015;100:1285–93.
Austin KM, Gupta ML Jr, Coats SA, Tulpule A, Mostoslavsky G, Balazs AB, et al. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008;118:1511–8.
Orelio C, Verkuijlen P, Geissler J, van den Berg TK, Kuijpers TW. SBDS expression and localization at the mitotic spindle in human myeloid progenitors. PLoS One. 2009;4:e7084.
Leung R, Cuddy K, Wang Y, Rommens J, Glogauer M. Sbds is required for Rac2-mediated monocyte migration and signaling downstream of RANK during osteoclastogenesis. Blood. 2011;117:2044–53.
Orelio C, Kuijpers TW. Shwachman-Diamond syndrome neutrophils have altered chemoattractant-induced F-actin polymerization and polarization characteristics. Haematologica. 2009;94:409–13.
In K, Zaini MA, Muller C, Warren AJ, von Lindern M, Calkhoven CF. Shwachman-Bodian-Diamond syndrome (SBDS) protein deficiency impairs translation re-initiation from C/EBPalpha and C/EBPbeta mRNAs. Nucleic Acids Res. 2016;44:4134–46.
Geiger H, Zheng Y. Regulation of hematopoietic stem cell aging by the small RhoGTPase Cdc42. Exp Cell Res. 2014;329:214–9.
Florian MC, Dorr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10:520–30.
Yang L, Zheng Y. Cdc42: a signal coordinator in hematopoietic stem cell maintenance. Cell Cycle. 2007;6:1445–50.
Mizukawa B, O’Brien E, Moreira DC, Wunderlich M, Hochstetler CL, Duan X, et al. The cell polarity determinant CDC42 controls division symmetry to block leukemia cell differentiation. Blood. 2017;130:1336–46.
van Hennik PB, de Koning AE, Ploemacher RE. Seeding efficiency of primitive human hematopoietic cells in nonobese diabetic/severe combined immune deficiency mice: implications for stem cell frequency assessment. Blood. 1999;94:3055–61.
Geiger H, True JM, de Haan G, Van Zant G. Age- and stage-specific regulation patterns in the hematopoietic stem cell hierarchy. Blood. 2001;98:2966–72.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. https://www.R-project.org/.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41.
Zhang X, Shang X, Guo F, Murphy K, Kirby M, Kelly P, et al. Defective homing is associated with altered Cdc42 activity in cells from patients with Fanconi anemia group A. Blood. 2008;112:1683–6.
Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci USA. 2007;104:5091–6.
Florian MC, Nattamai KJ, Dorr K, Marka G, Uberle B, Vas V, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503:392–6.
Guidi N, Sacma M, Standker L, Soller K, Marka G, Eiwen K, et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 2017;36:840–53.
Grigoryan A, Guidi N, Senger K, Liehr T, Soller K, Marka G, et al. LaminA/C regulates epigenetic and chromatin architecture changes upon aging of hematopoietic stem cells. Genome Biol. 2018;19:189.
Florian MC, Klose M, Sacma M, Jablanovic J, Knudson L, Nattamai KJ, et al. Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 2018;16:e2003389.
Liu W, Du W, Shang X, Wang L, Evelyn C, Florian MC, et al. Rational identification of a Cdc42 inhibitor presents a new regimen for long-term hematopoietic stem cell mobilization. Leuk: Off J Leuk Soc Am, Leuk Res Fund, UK. 2019;33:749–61.
Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–47.
Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–7.
Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–6.
Mizukawa B, O’Brien E, Moreira DC, Wunderlich M, Hochstetler CL, Duan X, et al. Cell polarity determinant CDC42 controls division symmetry to block leukemia cell differentiation. Blood. 2017;130:1336–46.
Bluteau O, Sebert M, Leblanc T, Peffault de Latour R, Quentin S, Lainey E, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131:717–32.
Warren AJ. Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul. 2018;67:109–27.
Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173:90–103.e19.
Acknowledgements
We thank Jeff Bailey and Victoria Summey from CCHMC Comprehensive Mouse and Cancer Core for their help with transplantation experiments and mouse bleed and the TTDSL at CCHMC for providing control BM MNCs samples. We thank Research Flow Cytometry Core at CCHMC for support with cell sorting and FACS analyzers. We thank the Schwachman–Diamond Syndrome registry for access to clinical data and samples. SK is currently supported by a Ramalingaswami fellowship, India at CSIR-Central Drug Research Institute, Lucknow. HG is supported by NIH grant DK104814. KCM is funded by a Conquer Cancer Foundation of ASCO Career Development Award, K12 HD028827. RNA-Seq data are archived (accession number will be provided upon publication).
Author information
Authors and Affiliations
Contributions
SK performed, analyzed experiments, interpreted data, prepared figures, and wrote the manuscript. KCM and HG designed and interpreted experiments, prepared figures, and wrote the manuscript. KJN, AH, AA, CZ, YL, and ML performed and analyzed experiments. RK and MM analyzed RNA-Seq data. AS, MCF, UB, AB, and SMD provided support with respect to experimental design.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kumar, S., Nattamai, K.J., Hassan, A. et al. Repolarization of HSC attenuates HSCs failure in Shwachman–Diamond syndrome. Leukemia 35, 1751–1762 (2021). https://doi.org/10.1038/s41375-020-01054-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-020-01054-8
This article is cited by
-
RNA binding protein SYNCRIP maintains proteostasis and self-renewal of hematopoietic stem and progenitor cells
Nature Communications (2023)
-
Longitudinal single-cell transcriptomics reveals distinct patterns of recurrence in acute myeloid leukemia
Molecular Cancer (2022)