Abstract
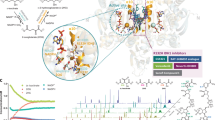
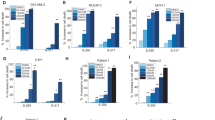
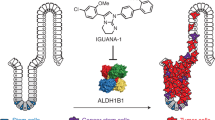
Mutations in isocitrate dehydrogenase 1 (IDH1) are found in 6% of AML patients. Mutant IDH produces R-2-hydroxyglutarate (R-2HG), which induces histone- and DNA-hypermethylation through the inhibition of epigenetic regulators, thus linking metabolism to tumorigenesis. Here we report the biochemical characterization, in vivo antileukemic effects, structural binding, and molecular mechanism of the inhibitor HMS-101, which inhibits the enzymatic activity of mutant IDH1 (IDH1mut). Treatment of IDH1mut primary AML cells reduced 2-hydroxyglutarate levels (2HG) and induced myeloid differentiation in vitro. Co-crystallization of HMS-101 and mutant IDH1 revealed that HMS-101 binds to the active site of IDH1mut in close proximity to the regulatory segment of the enzyme in contrast to other IDH1 inhibitors. HMS-101 also suppressed 2HG production, induced cellular differentiation and prolonged survival in a syngeneic mutant IDH1 mouse model and a patient-derived human AML xenograft model in vivo. Cells treated with HMS-101 showed a marked upregulation of the differentiation-associated transcription factors CEBPA and PU.1, and a decrease in cell cycle regulator cyclin A2. In addition, the compound attenuated histone hypermethylation. Together, HMS-101 is a unique inhibitor that binds to the active site of IDH1mut directly and is active in IDH1mut preclinical models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–64.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44.
Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–44.
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67.
Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Goparaju R, Schwarzer A, et al. Enantiomer-specific and paracrine leukemogenicity of mutant IDH metabolite 2-hydroxyglutarate. Leukemia. 2016;30:1708–15.
Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–57.
Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010;20:1188–200.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30.
Davis M, Pragani R, Popovici-Muller J, Gross S, Thorne N, Salituro F, et al. ML309: a potent inhibitor of R132H mutant IDH1 capable of reducing 2-hydroxyglutarate production in U87 MG glioblastoma cells. In: Probe Reports from the NIH Molecular Libraries Program. MD, USA: National Center for Biotechnology Information (US);2010. http://www.ncbi.nlm.nih.gov/pubmed/23905201.
Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878–86.
Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D, et al. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia. 2017;31:2020–8.
DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N Engl J Med. 2018;378:2386–98.
DiNardo CD, Schimmer AD, Yee KWL, Hochhaus A, Kraemer A, Carvajal RD, et al. A phase I study of IDH305 in patients with advanced malignancies including relapsed/refractory AML and MDS that harbor IDH1R132 mutations. Blood. 2016;128:1073–1073.
Watts JM, Baer MR, Lee S, Yang J, Dinner SN, Prebet T, et al. A phase 1 dose escalation study of the IDH1m inhibitor, FT-2102, in patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS). J Clin Oncol. 2018;36:7009–7009.
Pusch S, Krausert S, Fischer V, Balss J, Ott M, Schrimpf D, et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017;133:629–44.
Ma R, Yun CH. Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2. Biochem Biophys Res Commun. 2018;503:2912–7.
Xie X, Baird D, Bowen K, Capka V, Chen J, Chenail G, et al. Allosteric mutant IDH1 inhibitors reveal mechanisms for IDH1 mutant and isoform selectivity. Structure. 2017;25:506–13.
Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–87.
Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74.
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–21.
Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–42.
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf modeling. 2012;52:1757–68.
Urban DJ, Martinez NJ, Davis MI, Brimacombe KR, Cheff DM, Lee TD, et al. Assessing inhibitors of mutant isocitrate dehydrogenase using a suite of pre-clinical discovery assays. Sci Rep. 2017;7:12758.
Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–9.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30.
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83.
Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8.
Dang L, Su SM. Isocitrate dehydrogenase mutation and (R)-2-hydroxyglutarate: from basic discovery to therapeutics development. Annu Rev Biochem. 2017;86:305–31.
Deng G, Shen J, Yin M, McManus J, Mathieu M, Gee P, et al. Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J Biol Chem. 2015;290:762–74.
Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–71.
Acknowledgements
We acknowledge the assistance of the Cell Sorting Core Facility of Hannover Medical School supported in part by the Braukmann-Wittenberg-Herz-Stiftung and the Deutsche Forschungsgemeinschaft. We would like to thank all participating patients and contributing doctors, the staff of the Central Animal Facility of Hannover Medical School, and Silke Glowotz, Nadine Kattre, Girish Rajendraprasad, Roopsee Anand, Martin Wichmann, Petra Baruch, and Claudia Thiel for their support. This work was supported by an ERC grant under the European Union’s Horizon 2020 research and innovation program (No. 638035), by grant 70112697 from Deutsche Krebshilfe; and DFG grants HE 5240/5-1, HE 5240/5-2, HE 5240/6-1, and HE 5240/6-2.
Author information
Authors and Affiliations
Contributions
AC, RG, MP, and MH conceived and designed the study. AC, RG, CG, JW,TK, MM, AK, KG, RS, BO, ES, HB, D GK, and KB, collected the data. AC, RG, CG, MP, and MH analyzed and assembled the data. AG and MH collected patient samples and provided the patient data. AC, MP, and MH wrote the manuscript. All authors reviewed the data and edited and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
AC, MP, and MH have filed an EP and US patent application for HMS-101 (based on PCT/EP2014/059898 with priority of 2013). The remaining authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chaturvedi, A., Goparaju, R., Gupta, C. et al. In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site. Leukemia 34, 416–426 (2020). https://doi.org/10.1038/s41375-019-0582-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0582-x
This article is cited by
-
IDH1/2 mutations in acute myeloid leukemia patients and risk of coronary artery disease and cardiac dysfunction—a retrospective propensity score analysis
Leukemia (2021)
-
Safety and efficacy of BAY1436032 in IDH1-mutant AML: phase I study results
Leukemia (2020)
-
Combination treatment of an IDH1 inhibitor with chemotherapy in IDH1 mutant acute myeloid leukemia
Annals of Hematology (2020)