Abstract
BRCA1/BRCA2-containing complex 3 (BRCC3) is a Lysine 63-specific deubiquitinating enzyme (DUB) involved in inflammasome activity, interferon signaling, and DNA damage repair. Recurrent mutations in BRCC3 have been reported in myelodysplastic syndromes (MDS) but not in de novo AML. In one of our recent studies, we found BRCC3 mutations selectively in 9/191 (4.7%) cases with t(8;21)(q22;q22.1) AML but not in 160 cases of inv(16)(p13.1q22) AML. Clinically, AML patients with BRCC3 mutations had an excellent outcome with an event-free survival of 100%. Inactivation of BRCC3 by CRISPR/Cas9 resulted in improved proliferation in t(8;21)(q22;q22.1) positive AML cell lines and together with expression of AML1-ETO induced unlimited self-renewal in mouse hematopoietic progenitor cells in vitro. Mutations in BRCC3 abrogated its deubiquitinating activity on IFNAR1 resulting in an impaired interferon response and led to diminished inflammasome activity. In addition, BRCC3 inactivation increased release of several cytokines including G-CSF which enhanced proliferation of AML cell lines with t(8;21)(q22;q22.1). Cell lines and primary mouse cells with inactivation of BRCC3 had a higher sensitivity to doxorubicin due to an impaired DNA damage response providing a possible explanation for the favorable outcome of BRCC3 mutated AML patients.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is caused by genetic alterations that lead to enhanced proliferation and a differentiation block in hematopoietic progenitor cells. Although remissions can be achieved in the majority of patients with intensive chemotherapy, a large proportion of patients relapse which is associated with impaired survival. Recurrent genomic changes found in AML include structural chromosomal aberrations, translocations, and gene mutations. The most frequently found translocations/inversions are t(8;21)(q22;q22.1) and inv(16)(p13.1q22) or t(16;16)(p13.1q22) that together form the subgroup of core-binding factor (CBF) AML [1,2,3,4]. CBF is a heterodimeric transcription factor complex comprising RUNX1 and CBFB that is essential for the expression of genes vital for normal hematopoiesis. Lack or mutation of one of the complex members (RUNX1 in t(8;21)(q22;q22.1) AML or CBFB in inv(16)(p13.1q22) AML) results in a dominant negative inhibitory effect on CBF complex formation and impairment of hematopoiesis [5, 6]. The cytogenetic rearrangement t(8;21)(q22;q22.1) results in the fusion gene RUNX1-RUNX1T1 (AML1-ETO) [1, 2]. While the presence of AML1-ETO is considered to impair myeloid differentiation, mouse models suggest that it is not sufficient to induce leukemia [7]. Consistently, almost all AML cases with the t(8;21)(q22;q22.1) abnormality have concurrent genomic aberrations like mutations in KIT, FLT3, NRAS, and ASXL2 [8, 9]. Although they share similar biological effects like repression of CBF target genes and clinical characteristics including a favorable prognosis [10], t(8;21)(q22;q22.1) and inv(16)(p13.1q22) AML differ in regard to co-occurring mutations. For example, mutations in ASXL1 and ASXL2 occur at a high frequency in AML with t(8;21)(q22;q22.1) but are absent in AML with an inv(16)(p13.1q22) implying that they selectively cooperate with AML1-ETO [11,12,13].
BRCA1-/BRCA2-containing complex 3 (BRCC3, also known as BRCC36) is a Lysine-63-specific deubiquitinating enzyme (DUB) and member of the Zn2+ dependent JAB1/MPN/Mov34 metalloenzyme (JAMM) domain metalloprotease family [14, 15]. In contrast to ubiquitin chains linked at other another lysine residue, lysine-63 ubiquitination does in general not lead to proteasomal degradation of the target protein but instead to altered protein function. BRCC3 is a member of at least two complexes: the nuclear BRCA1-A complex consisting of Abraxas, BABAM1/MERIT40, BABAM2/BRCC45, UIMC1/RAP80, and BRCC3 [16,17,18], and the cytoplasmic BRCC36 isopeptidase complex (BRISC) including ABRO1, BABAM1, BABAM2, SHMT2, and BRCC3 [19,20,21,22,23]. Following DNA double strand breaks (DSBs), the E3 ubiquitin ligase RNF8 specifically adds K63-ubiquitin to histones H2A and H2AX [24], which in turn get recognized by the tandem ubiquitin interacting motif (UIM) of UIMC1 [25]. This leads UIMC1 to guide the BRCA1-A complex towards the DSB for DNA repair. BRCC3 finally resolves the interaction of the BRCA1-A complex with the DNA by cleaving off the K63-ubiquitin chains from H2A and H2AX [16]. Accordingly, mutations or deletions of BRCC3 have been shown to result in impaired DNA damage repair [26]. As a member of the BRISC, BRCC3 is involved in deubiquitination and thus regulation of different targeting proteins, including NLR family pyrin domain containing 3 (NLRP3) and type 1 interferon (IFN) receptor chain 1 (IFNAR1) [20, 27]. Loss of BRCC3 has been linked to decreased release of mature interleukin (IL)-1β by dysregulation of NLRP3 and the inflammasome [27] while impaired deubiquitination of IFNAR1 is associated with diminished induction of interferon signaling [20].
While recurrent mutations in BRCC3 have been recently described in myelodysplastic syndromes (MDS) [28], only one BRCC3 mutation has been identified in a patient with complex karyotype in the TCGA AML cohort of 200 cases [29]. In one of our sequencing studies of a large cohort of CBF AML patients, we exclusively found recurrent BRCC3 mutations in t(8;21)(q22;q22.1) AML [30]. Here, we investigated the impact of BRCC3 mutations on malignant transformation and drug sensitivity in human and murine hematopoietic cells.
Materials and methods
Patients
The targeted sequencing study included 351 CBF AML cases [30] with a t(8;21)(q22;q22.1) (n = 190) or inv(16)(p13.1q22) (n = 161). Genetic analyses were approved by the local ethics committee and all patients provided informed consent in accordance with the declaration of Helsinki. Further details about the patients and treatment are given in the supplementary information.
Plasmids
The following plasmids were previously described: pLKO5d.SFFV.SpCas9.P2A.BSD, pLKO5.sgRNA.EFS.PAC, pLKO5.hU6.sgRNA.dTom, and pRSF91-MCS-IRES-GFP-T2A-Puro [31]. pMY-IRES-GFP and pMY-IRES-GFP-AML1/ETO were kindly provided by F. Oswald (Internal Medicine I, University Hospital of Ulm). IFNAR1_pCSdest was a gift from Roger Reeves (Addgene plasmid # 53881) [32].
Genetic inactivation by CRISPR/Cas9 in cell lines
Kasumi-1, SKNO-1, THP-1, OCI-AML5, HEK-293T, and 32D cell lines stably expressing the Cas9 enzyme were generated using the pLKO5d.SFFV.SpCas9.P2A.BSD lentiviral vector and selected with blasticidin (10 µg/ml) (ant-bl, InvivoGen, San Diego, CA, USA). Presence of the FLAG-tagged Cas9 enzyme was confirmed by western blot. When generating a BRCC3 or Brcc3 knockout, vectors containing one or multiple sgRNA against BRCC3 or Brcc3 were introduced into the cell using the pLKO5.sgRNA.EFS.PAC plasmid and selected with puromycin (2 µg/ml) (ant-pr-1, InvivoGen). The following sgRNA-sequences were used: BRCC3 #1: AGCGTGGTTGAGACAAACG; BRCC3 #2: TCTAGTTGAACGATGATACA; Brcc3 #1: GGAGGTAAGTTGGCCACCT; Brcc3 #2: TGTGTATAGGGGAGGTAAGT; Brcc3 #3: TGTCATCATTCAACTAGAGT; Luciferase control: AGTTCACCGGCGTCATCGTC. The knockout was confirmed by western blot and Sanger sequencing.
Western blot
Protein concentrations were determined using the BCA Protein Assay Kit (23225, Thermo Fisher Scientific). The following antibodies were used: BRCC3: ab108411, Abcam, Cambridge, UK; Tubulin: T5168, Sigma-Aldrich; ABRO1: ab74333, Abcam; Abraxas: ab139191, Abcam; BABAM1: sc-160990, Santa Cruz, Dallas, TX, USA; UIMC1: 14466S, New England Biolabs, Ipswich, MA, USA; FLAG-tag (DYKDDDDK): 2368, Cell Signaling Technology, Danvers, MA, USA; HA-tag (YPYDVPDYA): 901533, BioLegend; G-CSF: sc-53292, Santa Cruz; β-Actin HRP: ab20272, Abcam; secondary anti-mouse HRP: 7076S, Cell Signaling; secondary anti-rabbit HRP: 7074S, Cell Signaling.
DNA damage and apoptosis
To assess DNA damage, Kasumi-1 BRCC3 WT, R81X, R81G, or KO cells were cultured at a density of 1 × 106 cells and treated with different concentrations of doxorubicin (S1208, SelleckChem, Houston, TX, USA) or DMSO as a vehicle control for 3 days. The cells were then fixated in ice-cold methanol overnight, stained with an anti-phospho histone H2A.X antibody (05-636-AF647, Merck Millipore) and analyzed using flow cytometry.
To measure the percentage of apoptotic cells in Kasumi-1 BRCC3 WT and KO cells, 1 × 106 cells were seeded and treated with different concentrations of doxorubicin or DMSO as a vehicle control for 3 days. Then, apoptosis was measured using the FITC Annexin V Apoptosis Detection Kit with 7-AAD (640922, BioLegend, San Diego, CA, USA) according to the manufacturer’s protocol.
Statistics
Differences between groups were analyzed by an unpaired t-test. For contingency tables, a Fisher’s exact test was applied. Kaplan–Meier plots were generated for each time-to-event outcome measure and differences between two groups were analyzed using the two-sided log-rank test. P values are displayed as follows: n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001. All statistical analyses were performed using Prism version 7 (GraphPad Software, San Diego, CA, USA).
A description of further methods is given in the Supplementary information.
Results
BRCC3 mutations in AML with t(8;21)(q22;q22.1)
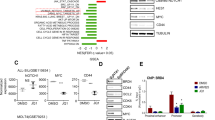
We first discovered BRCC3 mutations in two AML cases (Table 1, patients number #1 and #2) by exome sequencing (data unpublished). Since both BRCC3 mutated patients had a t(8;21)(q22;q22.1) and no BRCC3 mutations were found in other types of AML we included BRCC3 in a targeted sequencing study with a total of 351 patients with core-binding-factor AML including 191 patients with t(8;21)(q22;q22.1) and 160 patients with inv(16)(p13.1q22) [30]. Here, an additional seven BRCC3 mutations were found for a total of nine (4.7%) mutations in cases with t(8;21)(q22;q22.1) but none were found in the patient cohort with inv(16)(p13.1q22) (Fig. 1a) [30]. Three of the nine mutations were nonsense mutations, two frameshift mutations, three missense mutations and one mutation affected a splice site (Fig. 1b, Table 1). Seven of the nine mutations were located in the catalytically important MPN domain. Using the in silico prediction tool Polymorphism Phenotyping v2 (PolyPhen-2) algorithm [33], the coding mutations were all predicted to be damaging to the function of BRCC3. Four of the mutations affected a codon that was previously found to be mutated in MDS [28]. The BRCC3 gene is located on chromosome Xq28 and BRCC3 mutations were thus hemizygous in six male and two female patients with loss of one of the X chromosomes. However, based on the variant-allele fraction the majority of BRCC3 mutations were subclonal (Table 1).
BRCC3 mutation status in AML and MDS. a Frequency of BRCC3 mutations found in t(8;21)(q22;q22.1) and inv(16)(p13.1q22) AML in 351 patients [30]. Data for MDS and de novo AML was obtained from Huang et al. [28] and the TCGA AML [29] data set, respectively. b Overview of the BRCC3 coding region with mutations analyzed in t(8;21)(q22;q22.1) AML in our study on top and mutations found in Huang et al. and the TCGA data set below. The metalloprotease (MPN) domain is indicated in blue and the JAB1/MPN/Mov34 metalloenzyme (JAMM) domain in red
BRCC3 mutations are associated with a favorable prognosis in AML with t(8;21)(q22;q22.1)
Correlation with pretreatment clinical data including age, sex, leukocyte count, blood and bone marrow blast count, hemoglobin, platelet counts, or serum LDH levels revealed no significant differences between BRCC3 mutated and nonmutated patients (Table 2). Also, the number of additional cytogenetic alterations did not differ between BRCC3 mutated and nonmutated patients. We then investigated whether BRCC3 mutations affect response and outcome in patients with t(8;21)(q22;q22.1) treated with intensive chemotherapy (Fig. 2). All patients with BRCC3 mutations achieved a complete remission after induction therapy as compared with 87% of the patients without BRCC3 mutation (Fig. 2a). None of the patients with a BRCC3 mutation relapsed resulting in an event-free-survival and overall survival of 100% at 4 years (Fig. 2b, c).
Impact of BRCC3 mutations on clinical outcome. a Response after induction therapy in BRCC3 mutated and nonmutated patients. CR, complete remission; RD, refractory disease; ED, early death. Kaplan–Maier curves for event-free survival (b) and overall survival (c) in patients according to BRCC3 mutation status. Differences between groups were analyzed by a two-sided log-rank test
In order to determine whether the transcriptional level of BRCC3 has an impact on outcome in AML, we analyzed the TCGA de novo AML data set [29] with the BloodSpot online tool [34]. Here, patients with low BRCC3 levels had a better overall survival than patients with high BRCC3 levels (Supplementary Fig. S1).
Knockout of BRCC3 leads to increased cell proliferation in t(8;21)(q22;q22.1) AML cells
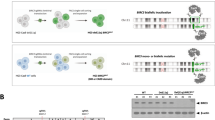
In order to analyze whether loss of BRCC3 affects cell growth, we determined the effect of CRISPR/Cas9-mediated BRCC3 inactivation in the two AML cell lines with t(8;21)(q22;q22.1) Kasumi-1 and SKNO-1 and in the OCI-AML5 cell line without t(8;21)(q22;q22.1). Complete knockout (KO) of BRCC3 was confirmed by western blot (Fig. 3a). We found that BRCC3 KO resulted in a significantly higher proliferation rate in the two AML cell lines with t(8;21)(q22;q22.1) Kasumi-1 and SKNO-1 (Fig. 3b). In contrast, no differences in proliferation were observed between the OCI-AML5 BRCC3 WT and KO cells (Fig. 3c). Next, we investigated whether the inactivating BRCC3 mutations R81X and R81G found in patients with t(8;21)(q22;q22.1) AML had a similar beneficial effect on cell proliferation in Kasumi-1. To this end, we ectopically over-expressed BRCC3 WT, R81X, or R81G in BRCC3 KO cells. Again, cell proliferation of Kasumi-1 BRCC3 R81X and R81G expressing cells was significantly higher as compared with BRCC3 WT cells suggesting that both mutants are inactivating (Fig. 3d). Lastly, BRCC3 has been found mutated at a subclonal level in some t(8;21)(q22;q22.1) AML patients. We therefore analyzed the impact of a subclone of BRCC3 KO on cell proliferation. We found that a fraction of 40% of Kasumi-1 cells harboring a BRCC3 KO was sufficient to significantly increase cell proliferation of all cells in the culture as compared with BRCC3 WT cells (Fig. 3e). Of note, the number of cells harboring the BRCC3 KO remained constant at ~40% implying that the BRCC3 KO cells stimulated proliferation of BRCC3 KO and WT cells (Supplementary Fig. S2).
Impact of BRCC3 mutations on proliferation and self-renewal in vitro. a Validation of BRCC3 knockout via CRISPR/Cas9 in Kasumi-1, SKNO-1, and OCI-AML5 cell lines using western blot. Growth curve of Kasumi-1, SKNO-1, (b) and OCI-AML5 (c) cells containing either BRCC3 wild type (WT) or knockout (KO) for a duration of 14 or 18 days. d Growth curve of BRCC3 R81X and R81G mutations in Kasumi-1 cells compared with BRCC3 WT cells over a duration of 11 days. e Growth curve of BRCC3 KO cells at a subclonal level (40% BRCC3 KO + 60% BRCC3 WT) in Kasumi-1 cells over a duration of 12 days. f Schematic overview of colony forming unit (CFU) assay using sorted murine hematopoietic stem- and progenitor cells (LSK cells) infected with a retrovirus expressing AML1-ETO or a control and a lentivirus expressing a pool of three different sgRNAs targeting Brcc3 or a non-targeting sgRNA control. After infection, CFUs were seeded for 10–14 days and serially replated. g Colony forming unit (CFU) assay. h Microscopic assessment of cells taken from the 8th replate of AML1-ETO + Brcc3 KO. Original magnification ×100. Significance was determined using unpaired Student's t test. nsNot significant, *P < 0.05, **P < 0.01, ***P < 0.001. n = 3 for data shown, which are representative of three independent experiments
In the next step, we investigated whether loss of Brcc3/BRCC3 is capable of conferring cytokine-independence in murine myeloid 32D or SKNO-1 cells. We found that Brcc3-inactivation by CRISPR/Cas9 did not convey IL-3-independent growth in 32D cells as compared with KrasG13D expression that was used as a positive control (Supplementary Fig. S3a). Likewise, loss of BRCC3 did not render SKNO-1 cells independent of GM-CSF (Supplementary Fig. S3b). Similarly, inactivation of Brcc3/BRCC3 did not confer cytokine-independence to Ba/F3 and TF-1 cells (data not shown).
Loss of Brcc3 in combination with AML1-ETO leads to unlimited self-renewal capacity
In order to investigate the effect of Brcc3 inactivation on the self-renewal capacity in mouse hematopoietic progenitor cells we performed colony forming assays. To this end, we infected primary murine stem- and progenitor Lineage- Sca1+ c-Kit+ (LSK) cells isolated from the transgenic Rosa26-Cas9 mouse that constitutively express the Cas9 enzyme [35] with retroviral vectors containing either AML1-ETO or a control as well as a lentiviral vector expressing sgRNAs targeting Brcc3 or a non-targeting sgRNA control (Fig. 3f). While the colony number of cells containing either a control, a single knockout of Brcc3, or AML1-ETO alone decreased rapidly after 3–5 replatings, cells expressing both, AML1-ETO and the Brcc3-specific sgRNAs could be replated at least eight times (Fig. 3g). Microscopic assessment of the cells demonstrated an immature blast population in cells transduced with AML1-ETO + Brcc3KO (Fig. 3h) while in the other conditions most cells had a more differentiated phenotype (Supplementary Fig. S4).
Mutations in BRCC3 lead to impaired interleukin and interferon signaling
To further analyze the functional impact of BRCC3 mutations found in AML t(8;21)(q22;q22.1) and MDS patients, we cloned the two missense mutations R81G and R82H, located within the MPN + domain as well as D135E located at the end of the JAMM motif (Fig. 1b). When expressed in HEK-293T cells, all three mutations maintained structural integrity with other BRCA-1A and BRISC complex members at similar levels compared with wild type BRCC3 in a pull-down experiment (Supplementary Fig. S5a). However, both scaffolding proteins Abraxas and ABRO1 were downregulated in Kasumi-1 BRCC3 KO cells while UIMC1 and BABAM1 were not affected (Supplementary Fig. S5b). In a stable isotope labeling of amino acids in cells (SILAC)-based proteomic interaction partner analysis of FLAG-tagged BRCC3 WT and two recurrent point mutants (R81G and D135E) we did not observe relevant changes in binding to known BRCC3 interactors (Supplementary Fig. S6a–c).
Previous studies have identified IFNAR1 and NLRP3 as substrates of the BRCC3 isopeptidase complex (BRISC) [20, 27]. Both proteins rely on deubiquitination mediated by BRCC3 to exercise their cellular functions and shRNA/siRNA mediated knockdown of BRCC3 has been implicated in impaired activation of IFNAR1 and NLRP3 [20, 27]. NLRP3 is an essential member of the inflammasome that regulates IL-1β. In stimulated THP-1 we found that inactivation of BRCC3 led to significantly diminished IL-1β release (Supplementary Fig. S7a). We then went on to analyze whether addition of IL-1β has a negative effect on proliferation of BRCC3 WT and KO Kasumi-1 and SKNO-1 cells. While SKNO-1 BRCC3 WT had a moderately enhanced proliferation in the presence of exogenous IL-1β (Supplementary Fig. S7b), neither SKNO-1 BRCC3 KO nor Kasumi-1 BRCC3 WT or KO showed differences in proliferation (Supplementary Fig. S7b, c). Next, we investigated the effect of BRCC3 mutants on IFNAR1 ubiquitination in BRCC3 KO HEK-293T cells. K63-linked ubiquitination levels of IFNAR1 were higher in cells expressing BRCC3 mutants as compared with BRCC3 WT expressing cells (Fig. 4a). When we checked for interferon response-genes depending on IFNAR1 in Kasumi-1 cells, we observed that upon stimulation with IFN, the induction of IFN response genes IFI44 (Fig. 4b) as well as of OASL (Supplementary Fig. S7d) were diminished in BRCC3 knockout cells. IFNα has been shown to decrease self-renewal capacity of cells with a t(8;21)(q22;q22.1) background [36]. Consistently, IFNα treatment impaired proliferation of Kasumi and SKNO-1 cells and this effect could be rescued in part by inactivation of BRCC3 (Fig. 4c).
Influence of BRCC3 mutations on IFNAR1 ubiquitination and IFN signaling. a Ubiquitination analysis of IFNAR1 in HEK-293T BRCC3 KO cells expressing wild-type or mutant BRCC3. b mRNA expression of the IFN-response gene IFI44 in Kasumi-1 cells with or without treatment with interferon. c Growth curves of BRCC3 WT or BRCC3 KO Kasumi-1 and SKNO-1 cells treated with 10 ng/mL IFNα or a vehicle control for a duration of 11 days. nsNot significant, *P < 0.05, **P < 0.01, ***P < 0.001. All p-values are calculated using unpaired Student t test. Error bars are standard error of the mean. n = 2 for Fig. 4a, n = 3 for all other experiments, which are representative of two or three independent experiments respectively
Loss of BRCC3 induces increased cytokine signaling
Given the role of BRCC3 in regulation of several cytokines like IL-1β and IFN, we looked for the impact of BRCC3 inactivation on cytokine release systematically using a cytokine array in unstimulated Kasumi-1 cells. This revealed an upregulation of several cytokines in BRCC3 KO cells with G-CSF (1.75 ± 0.03) being the top hit (Fig. 5a). Upregulation of G-CSF release was also confirmed by western blot (Fig. 5b). Given the growth stimulating effects of G-CSF on certain AML subtypes including t(8;21)(q22;q22.1) we treated several cell lines with G-CSF. This revealed that cell proliferation of both t(8;21)(q22;q22.1) AML cell lines Kasumi-1 and SKNO-1 with intact BRCC3 significantly increased when treated with G-CSF (Fig. 5c, d). In contrast, G-CSF did not further enhance the growth of BRCC3 KO Kasumi-1 cells (Fig. 5e). G-CSF also had no impact on cell proliferation in non-t(8;21)(q22;q22.1) AML cell lines THP (Supplementary Fig. S8) or the inv(16)(p13.1q22) AML cell line ME-1 (Fig. 5f). When treated with a neutralizing anti-GCSF antibody, cell proliferation of Kasumi-1 BRCC3 KO but not BRCC3 WT cells decreased (Fig. 5g). Finally, G-CSF increased the colony number of LSK cells transduced with AML1-ETO to numbers comparable to cells transduced with AML1-ETO + Brcc3KO which could not be further stimulated (Fig. 5h).
Impact of BRCC3 inactivation on cytokine release. a Top up-regulated cytokines in Kasumi-1 BRCC3 KO supernatant compared to BRCC3 WT assessed via Cytokine Array. b Western blot depicting G-CSF levels in Kasumi-1 BRCC3 WT and BRCC3 KO cells. Growth curves of c BRCC3 WT Kasumi-1 and d SKNO-1, e BRCC3 KO Kasumi-1, and f BRCC3 WT ME-1 cells treated with 10 ng/mL G-CSF or a vehicle control for 8 days. g Kasumi-1 BRCC3 WT and BRCC3 KO cells treated with a neutralizing anti-G-CSF antibody for 13 days. h Colony forming unit (CFU) assay of AML1-ETO and AML1-ETO + Brcc3 KO cells treated with G-CSF or a vehicle control. nsNot significant, *P < 0.05, **P < 0.01, ***P < 0.001. All p-values are calculated using unpaired Student t test. Error bars are standard error of the mean. n = 2 for Fig. 5h, n = 3 for all other experiments for data shown, which are representative of two or three independent experiments respectively
Inactivation of BRCC3 results in enhanced sensitivity towards doxorubicin
Downregulation of BRCC3 has been associated with increased apoptosis in breast cancer cells following ionizing radiation and enhanced sensitivity towards temozolomide in human glioma cells [26, 37]. We investigated whether loss of BRCC3 renders Kasumi-1 cells more sensitive towards doxorubicin and cytarabine that are the standard treatment for AML with t(8;21)(q22;q22.1). For this purpose, either parental or CRISPR-mediated stable BRCC3 KO Kasumi-1 cells (Fig. 6a) were treated with doxorubicin for 7 days. We found that knockout with two different sgRNAs targeting BRCC3 led to a significant decrease in cell viability compared with cells transduced with control sgRNAs (Fig. 6b). Next, the two BRCC3 mutations R81X and R81G were ectopically over-expressed in BRCC3 KO cells. Like BRCC3 KO cells, both BRCC3 mutants also displayed an enhanced sensitivity towards doxorubicin (Fig. 6c). This enhanced sensitivity to doxorubicin was abrogated when we reintroduced BRCC3 WT by retroviral expression in BRCC3 KO cells (Fig. 6d). In contrast to doxorubicin, the sensitivity towards cytarabine was not affected (Fig. 6b). When we checked for DNA damage accumulation caused by doxorubicin treatment, we found that BRCC3 KO as well as BRCC3 R81X and R81G mutated cells accumulated significantly more DNA damage as measured by the level of phosphorylated γH2A.X (Fig. 6e). Similarly, the percentage of apoptotic cells was also significantly higher in BRCC3 KO cells post doxorubicin treatment (Fig. 6f). Lastly, we analyzed mouse LSK cells infected with either AML1-ETO or an empty control vector as well as sgRNAs targeting Brcc3 or a non-targeting sgRNA (Fig. 6g). Again, we observed that cells with Brcc3 inactivation were more sensitive to doxorubicin treatment as compared to cells with normal Brcc3 status.
Impact of BRCC3 inactivation on doxorubicin sensitivity. a Confirmation of CRISPR/Cas9 mediated knockout (KO) of BRCC3 for two different sgRNAs in Kasumi-1 cells by western blot. b Cell viability of parental or BRCC3 KO Kasumi-1 cells after treatment with 7.5 nM doxorubicin or 100 nM cytarabine for 7 days. c Cell viability of parental or BRCC3 R81X and R81G Kasumi-1 cells after treatment with 5 nM doxorubicin. d Cell viability of parental, BRCC3 KO, or BRCC3 KO ectopically expressing BRCC3 Kasumi-1 cells after treatment with 5 nM doxorubicin for 7 days. e Assessment of phosphorylated γ-H2A.X after treatment with 100 nM or 200 nM doxorubicin or a vehicle control for 3 days in BRCC3 WT, BRCC3 R81X, R81G, and KO cells using flow-cytometry. f Evaluation of apoptosis in Kasumi-1 BRCC3 WT or KO cells treated with 100 nM, 200 nM, or 300 nM doxorubicin or a vehicle control for a duration of 3 days. g Determination of sensitivity towards doxorubicin and cytarabine in sorted murine hematopoietic stem- and progenitor cells (LSK cells) infected with a retrovirus expressing AML1-ETO or empty control and a lentivirus expressing sgRNAs targeting Brcc3 or a non-targeting sgRNA control. nsNot significant, *P < 0.05, **P < 0.01, ***P < 0.001. All p-values are calculated using unpaired Student t test. Error bars are standard error of the mean. n = 3 for data shown, which is representative of three independent experiments
Discussion
Here, we investigated the role of BRCC3 mutations in the biology and treatment of t(8;21)(q22;q22.1) AML. In a large sequencing study of CBF AML we found recurrent inactivating mutations in BRCC3 in t(8;21)(q22;q22.1) AML with a frequency of 4.7% but not in inv(16)(p13.1q22) AML [30]. Consistently, BRCC3 mutations were found in 5 patients in a recent study of 331 t(8;21)(q22;q22.1) AML cases [38], but in only one patient with MDS-related cytogenetic abnormalities in the TCGA de novo AML cohort [29]. These results indicate a certain selectivity of BRCC3 mutations for t(8;21)(q22;q22.1) AML. In our cohort, BRCC3 mutations had no impact on preclinical characteristics but were associated with a favorable outcome with the caveat that the case number is small. Seven out of the nine BRCC3 mutations were located within the catalytically active MPN + domain, four of them at the exact position as mutations previously found in MDS, while the other three were located in close proximity [28]. Given that most BRCC3 mutations found in AML and MDS are predicted to be deleterious and are hemizygous we hypothesized that loss of BRCC3 function contributes to malignant transformation of hematopoietic cells in a specific genetic context.
In accordance with the close association of BRCC3 mutations with t(8;21)(q22;q22.1) we found a pro-proliferative effect of BRCC3 inactivation in the t(8;21)(q22;q22.1) AML cell lines Kasumi-1 and SKNO-1 but not in a cell line without t(8;21)(q22;q22.1). CRISPR-mediated BRCC3 inactivation alone did not enhance the self-renewal capacity in mouse hematopoietic progenitor cells (LSK) implying that loss of BRCC3 alone is not sufficient for malignant transformation. In contrast, self-renewal capacity was markedly increased in LSK cells when BRCC3 was inactivated together with expression of AML1-ETO supporting that both genetic alterations cooperate.
We next sought to investigate how inactivation of BRCC3 contributes to malignant transformation. One possible mechanism would be that loss of BRCC3 in the BRCA1-A complex leads to impaired DNA damage repair what may increase the likelihood for acquisition of genomic aberrations that activate oncogenic pathways. However, we considered this possibility for several reasons unlikely. First, in our AML t(8;21)(q22;q22.1) cohort no increase in secondary cytogenetic aberrations was observed and in MDS, BRCC3 mutations are associated with a normal karyotype [28]. Second, the majority of the BRCC3 mutations found in AML and also in MDS are subclonal, suggesting that they are a secondary event and not an initiating founder mutation that promotes further genetic alterations. Besides the role in DNA damage repair, BRCC3 is involved in other cellular pathways including cytokine signaling and regulation. BRCC3 as part of the BRISC-complex deubiquitinates its substrate IFNAR1 which affects interferon signaling [20]. We found that BRCC3 mutations abrogate the DUB activity on IFNAR1 resulting in an attenuated interferon response in AML cells. In a study by DeKelver et al. it was shown that AML1-ETO induces strong interferon signaling and that this antagonizes the leukemic potential of AML1-ETO [36]. While IFNAR1 has not been found mutated or deleted in t(8;21)(q22;q22.1) AML, we could show that BRCC3 mutations render t(8;21)(q22;q22.1) AML cells less sensitive to IFNα-mediated toxicity through impaired deubiquitination and function of IFNAR1. This provides a possible explanation for the close association of BRCC3 mutations with t(8;21)(q22;q22.1). We also demonstrate that AML cells with BRCC3 inactivation had an impaired inflammasome activity with decreased IL-1β release after activation. In our models, IL-1β had no negative effect on cell proliferation of AML1-ETO cell lines. However, inflammasome-driven IL-1β has been found to suppress AML proliferation in other models [39]. Thus, downregulation of IL-1β by defective BRCC3 deubiquitination of NLRP3 could contribute to the transforming and proproliferative effects of BRCC3 mutations.
In addition to IL-1β and interferon signaling, we found that BRCC3 inactivation leads to deregulation of release of other cytokines by AML cells. Although not sufficient to induce cytokine-independence in several cell lines, some of the more released cytokines in BRCC3 inactivated AML cells like G-CSF, HGF, and IL-4 have been shown to stimulate proliferation of AML cells [40,41,42,43]. We were able to directly tie increased G-CSF release to enhanced cell proliferation of t(8;21)(q22;q22.1) AML cells harboring BRCC3 WT, but not to other cell lines with a different cellular background including inv(16)(p13.1q22), which further links BRCC3 mutations to t(8;21)(q22;q22.1) AML. As loss of BRCC3 on a subclonal level was sufficient to increase cell proliferation, this implies that BRCC3 mutations at least in part act through a paracrine mechanism by release of G-CSF. A paracrine activation of AML cells would also explain how subclonal BRCC3 mutations, as present in some of the AML and MDS patients, stimulate proliferation of nonmutated clones through pro-proliferative cytokines. Remarkably, primary AML cells and cell lines with t(8;21)(q22;q22.1) have a higher expression of G-CSF receptor molecules (G-CSFR) than other AML subtypes [41, 44]. G-CSF induces tyrosine phosphorylation of JAK2 through G-CSFR which leads to enhanced proliferation [45]. Of note, in a very recent study by Donaghy et al. BRCC3-mediated deubiquitination of JAK2 has been implicated in limiting hematopoietic stem cell expansion and knockdown of BRCC3 was associated with an increased K63-ubiquitination and activation of JAK2 [46]. Therefore, BRCC3 inactivation enhances JAK2 signaling by two mechanisms. Remarkably, JAK2 signaling is a particularly important driver in t(8;21)(q22;q22.1) AML as indicated by the presence of activating JAK2 mutations specifically in this but not other AML subtypes [47].
Enhanced self-renewal and proliferation by BRCC3 mutations would suggest that they are associated with a more aggressive disease. However, the BRCC3 mutated t(8;21)(q22;q22.1) AML patients in our cohort had an excellent outcome. In addition, in the TCGA dataset of de novo AML lower BRCC3 expression was associated with a better overall survival [29]. Our observations in cell lines and murine primary hematopoietic cells suggest that BRCC3 inactivation leads to an impaired capability of the BRCA1-A complex to repair DNA damage and subsequently higher sensitivity to DNA damaging chemotherapy. In fact, none of the BRCC3 mutated patients in our cohort that received intensive chemotherapy relapsed. Consistently, downregulation of BRCC3 and its complex members has been previously associated with a higher sensitivity to DNA damage inducing agents in other types of cancer [26, 37].
In conclusion, we demonstrate that BRCC3 mutations lead to altered ubiquitination of its substrates and cytokine release which cooperates with AML1-ETO to induce AML and sensitizes the leukemic cells to cytotoxic chemotherapy. Future studies are needed to investigate the clinical impact of BRCC3 mutations in larger cohorts in the context of different treatments and elucidate whether altered ubiquitination of additional, yet unknown substrates of BRCC3 contributes to the development of AML and MDS.
References
Hagemeijer A, Garson OM, Kondo K. Fourth international workshop on chromosomes in leukemia 1982: translocation (8;21)(q22; q22) in acute nonlymphocytic leukemia. Cancer Genet Cytogenet. 1984;11:284–7.
Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–4.
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. New Engl J Med. 2015;373:1136–52.
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. New Engl J Med. 2016;374:2209–21.
Renneville A, Roumier C, Biggio V, Nibourel O, Boissel N, Fenaux P, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–31.
Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–13.
Downing JR. The core-binding factor leukemias: lessons learned from murine models. Curr Opin Genet Dev. 2003;13:48–54.
Kuhn MWM, Radtke I, Bullinger L, Goorha S, Cheng J, Edelmann J, et al. High-resolution genomic profiling of adult and pediatric core-binding factor acute myeloid leukemia reveals new recurrent genomic alterations. Blood. 2012;119:e67–75.
Micol J-B, Duployez N, Boissel N, Petit A, Geffroy S, Nibourel O, et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood. 2014;124:1445–9.
Schlenk RF, Benner A, Krauter J, Büchner T, Sauerland C, Ehninger G, et al. Individual patient data–based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2004;22:3741–50.
Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127:2451–9.
Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–6.
Jahn N, Agrawal M, Bullinger L, Weber D, Corbacioglu A, Gaidzik VI, et al. Incidence and prognostic impact of ASXL2 mutations in adult acute myeloid leukemia patients with t(8;21)(q22; q22): a study of the German–Austrian AML Study Group. Leukemia. 2017;31:1012–5.
Cope GA. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11.
Dong Y, Hakimi M-A, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–99.
Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci. 2009;106:3166–71.
Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202.
Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–54.
Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–31.
Zheng H, Gupta V, Patterson-Fortin J, Bhattacharya S, Katlinski K, Wu J, et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013;5:180–93.
Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem. 2010;285:30971–81.
Cooper EM, Boeke JD, Cohen RE. Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to Lys63-linked polyubiquitin. J Biol Chem. 2010;285:10344–52.
Feng L, Wang J, Chen J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem. 2010;285:30982–8.
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40.
Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5.
Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. BRCC36 is essential for ionizing radiation–induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66:5039–46.
Py BF, Kim M-S, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–8.
Huang D, Nagata Y, Grossmann V, Radivoyevitch T, Okuno Y, Nagae G, et al. BRCC3 mutations in myeloid neoplasms. Haematologica. 2015; http://www.haematologica.org/cgi/doi/10.3324/haematol.2014.111989
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Engl J Med. 2013;368:2059–74.
Jahn N, Agrawal M, Dolnik A, Cocciardi S, Schmalbrock LK, Blatte TJ, et al. Genetic heterogeneity of t(8;21)(q22; q22.1) acute myeloid leukemia revealed by high-throughput targeted sequencing. Blood. 2017;130(Suppl 1):2688.
Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–6.
Edie S, Zaghloul NA, Leitch CC, Klinedinst DK, Lebron J, Thole JF, et al. Survey of human chromosome 21 gene expression effects on early development in Danio rerio. 2018;8:2215–23.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7: Unit 7.20.
Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sønderby CK, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44(D1):D917–24.
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55.
DeKelver RC, Lewin B, Weng S, Yan M, Biggs J, Zhang D-E. RUNX1–ETO induces a type I interferon response which negatively effects t(8;21)-induced increased self-renewal and leukemia development. Leuk Lymphoma. 2014;55:884–91.
Chai KM, Wang C-Y, Liaw H-J, Fang K-M, Yang C-S, Tzeng S-F. Downregulation of BRCA1-BRCA2-containing complex subunit 3 sensitizes glioma cells to temozolomide. Oncotarget. 2014;5:10901–15.
Christen F, Hoyer K, Yoshida K, Hou H-A, Waldhueter N, Heuser M, et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133:1140–51.
Höckendorf U, Yabal M, Herold T, Munkhbaatar E, Rott S, Jilg S, et al. RIPK3 restricts myeloid leukemogenesis by promoting cell death and differentiation of leukemia initiating cells. Cancer Cell. 2016;30:75–91.
Guo J-R, Li W, Wu Y, Wu L-Q, Li X, Guo Y-F, et al. Hepatocyte growth factor promotes proliferation, invasion, and metastasis of myeloid leukemia cells through PI3K-AKT and MAPK/ERK signaling pathway. Am J Transl Res. 2016;8:3630–44.
Shimizu K, Kitabayashi I, Kamada N, Abe T, Maseki N, Suzukawa K, et al. AML1-MTG8 leukemic protein induces the expression of granulocyte colony-stimulating factor (G-CSF) receptor through the up-regulation of CCAAT/enhancer binding protein epsilon. Blood. 2000;96:288–96.
Rossi FM, Degan M, Mazzocco FT, Di Francia R, Aldinucci D, Poletto D, et al. Co-expression of CD30 ligand and interleukin 4 (IL-4) receptors by acute myeloid leukaemia blasts is associated with the expansion of IL-4-producing CD30+ normal T cells. Br J Haematol. 2002;117:59–69.
Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–22.
Motoji T, Watanabe M, Uzumaki H, Kusaka M, Fukamachi H, Shimosaka A, et al. Granulocyte colony-stimulating factor (G-CSF) receptors on acute myeloblastic leukaemia cells and their relationship with the proliferative response to G-CSF in clonogenic assay. Br J Haematol. 1991;77:54–9.
Shimoda K, Iwasaki H, Okamura S, Ohno Y, Kubota A, Arima F, et al. G-CSF induces tyrosine phosphorylation of the JAK2 protein in the human myeloid G-CSF responsive and proliferative cells, but not in mature neutrophils. Biochem Biophys Res Commun. 1994;203:922–8.
Donaghy R, Han X, Rozenova K, Lv K, Jiang Q, Doepner M, et al. The BRISC deubiquitinating enzyme complex limits hematopoietic stem cell expansion by regulating JAK2 K63-ubiquitination. Blood. 2019;133:1560–71.
Iwanaga E, Nanri T, Matsuno N, Kawakita T, Mitsuya H, Asou N. A JAK2-V617F activating mutation in addition to KIT and FLT3 mutations is associated with clinical outcome in patients with t(8;21)(q22;q22) acute myeloid leukemia. Haematologica. 2009;94:433–5.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (SFB-1074 to JK, KD, LB, HD, SW; and Emmy-Noether Program KR3886/2-1 to JK). TM and LR were supported by the Graduate School of Ulm, which is funded by the Excellence Initiative of the German Federal and State Governments. DH was supported by the Deutsche Krebshilfe (111743). We would like to thank Hartmut Geiger, Franz Oswald and the other members of SFB-1074 for fruitful discussions and comments and Franz Oswald for providing the pMY-IRES-GFP and pMY-IRES-GFP-AML1/ETO plasmids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Meyer, T., Jahn, N., Lindner, S. et al. Functional characterization of BRCC3 mutations in acute myeloid leukemia with t(8;21)(q22;q22.1). Leukemia 34, 404–415 (2020). https://doi.org/10.1038/s41375-019-0578-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-019-0578-6
This article is cited by
-
Deubiquitinase BRCC3 promotes the migration, invasion and EMT progression of colon adenocarcinoma by stabilizing MET expression
Genes & Genomics (2024)
-
DNA methylation landscape reveals LIN7A as a decitabine-responsive marker in patients with t(8;21) acute myeloid leukemia
Clinical Epigenetics (2023)
-
Deubiquitinases in hematological malignancies
Biomarker Research (2021)
-
AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms
Oncogene (2021)
-
HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells
Nature Communications (2020)