Abstract
Gastric cancer (GC) is one of the most common and deadly cancers in the world. It is a multifactorial disease highly influenced by environmental factors, which include radiation, smoking, diet, and infectious pathogens. Accumulating evidence suggests that epigenetic regulators are frequently altered in GC, playing critical roles in gastric tumorigenesis. Epigenetic regulation involves DNA methylation, histone modification, and noncoding RNAs. While it is known that environmental factors cause widespread alterations in DNA methylation, promoting carcinogenesis, the chromatin- and noncoding RNA-mediated mechanisms of gastric tumorigenesis are still poorly understood. In this review, we focus on discussing recent discoveries addressing the roles of histone modifiers and noncoding RNAs and the mechanisms of their interactions in gastric tumorigenesis. A better understanding of epigenetic regulation would likely facilitate the development of novel therapeutic approaches targeting specific epigenetic regulators in GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is the fifth most common cancer and the fourth deadliest cancer in the world1. GC shows distinct incidence patterns across geographical regions: most gastric cancer cases are found in Eastern Asia and Europe, while it is the most common cancer in some of Middle Eastern countries. Although the overall incidence and mortality have decreased in most countries, likely due to better prevention and improved food preservation, increasing incidence rates among young adult populations have been observed in some countries, such as the UK and Sweden2. Notably, the treatment of advanced or metastatic GC, with a median overall survival of less than one year, has been extremely challenging3. Therefore, a better understanding of the disease mechanisms would likely help design more effective therapies.
Consistent with its distinct geographical distribution patterns, GC is a highly multifactorial disease regulated by genetic, epigenetic, and environmental factors. Common genetic factors include mutations in oncogenes and tumor suppressor genes involved in cancer initiation and progression4. The Cancer Genome Atlas (TCGA) project has identified common mutations and categorized GC into four major subtypes: Epstein‒Barr virus (EBV)-positive, microsatellite instability (MSI), genomically stable (GS), and chromosomal instability (CIN)5. Epigenetic factors are involved in the regulation of gene expression via mechanisms unrelated to the genetic sequence, which include DNA methylation, histone modification, and noncoding RNAs (ncRNAs)6. Interestingly, analysis of tumor suppressors and oncogenes has shown that changes in DNA methylation are more common than genetic mutations in GC, highlighting the importance of epigenetic regulation in gastric tumorigenesis7. Corroborating these data, another study showed that the impact of DNA methylation outweighs that of genetic mutations in GC when compared to esophageal cancer8. Similarly, environmental factors, most notably diet and infection, play major roles in gastric carcinogenesis. In fact, GC is strongly associated with two infectious agents categorized as type I carcinogens, H. pylori and EBV9,10,11,12,13.
Environmental and epigenetic factors are interconnected, as both diet and infection have been found to influence DNA methylation14,15. While DNA methylation in GC has been extensively reviewed6,16,17,18, other types of epigenetic regulation in GC are still poorly understood. As newly accumulating evidence suggests the critical roles of histone modification and ncRNAs in GC tumorigenesis, we discuss recent studies demonstrating their roles and mechanistic interactions.
Histone modification and the chromatin landscape in gastric cancer
DNA is wrapped around octomeric histone cores to form nucleosomes, which are organized into chromatin. The organization of chromatin into accessible/active euchromatin or condensed/repressed heterochromatin directly influences gene expression, and it is tightly regulated by covalent modifications of the histone tails via methylation, acetylation and phosphorylation19. These modifications are reversible and are catalyzed by specialized “writer” and “eraser” proteins. For example, writers such as histone methyltransferases (HMTs) and histone acetyltransferases (HATs) deposit methyl and acetyl marks on histone tails, respectively. In contrast, erasers such as histone demethylases and histone deacetylases (HDACs) remove the respective marks20. Other types of modifications, such as ubiquitination, sumoylation, and GlcNAcylation for histones, are also possible, but they are much less studied. Writers and erasers also have specificity for the histone marks they regulate. For example, the catalytic subunit of the polycomb repressive complex 2 (PRC2), EZH2, catalyzes the methylation of histone H3 lysine 27 (H3K27), while H3K4 methylation is catalyzed by the complex proteins associated with the Set1 (COMPASS) complex. Similarly, histone lysine demethylase (KDM)6A/B/C removes methylation on H3K27, while LSD1/KDM1A demethylates H3K4me20.
The combination of histone modifications represents a code correlated with functional elements on the chromatin. For example, H3K4 methylation is associated with active elements: H3K4me3 and H3K4me1 are associated with enhancers and promoters, respectively21,22. Histone modifications can influence gene expression. For example, H3K27ac and H3K9ac are associated with active gene expression, while H3K27me3 is associated with transcriptional repression. The specific methylation state of the same amino acid residue can also function differently: H3K9me1 is associated with active promoters, while H3K9me2/3 is associated with promoter repression23.
Once histone marks are established, they can be recognized by “readers”, which are proteins that can interact with specific histone marks and exert a myriad of effects20. In this review, we will focus on ATPase-dependent chromatin remodeling complexes, which contain reader subunits and can directly influence the chromatin state by modifying either accessibility or nucleosome composition24,25.
Alterations of histone modifiers establish the histone code underlying gastric carcinogenesis
Together, writer and eraser proteins establish the histone code, influencing the chromatin landscape and regulating a multitude of signaling pathways and genes. Unsurprisingly, many of these proteins play important roles in oncogenesis and tumor suppression. Since dysregulation of writer and eraser proteins in GC has been previously discussed17,26,27, here, we focus on the recent discoveries addressing their roles in GC.
Histone methyltransferases
Analysis of TCGA data showed that a number of histone modifiers are differentially expressed between GC and noncancer samples28. A more recent analysis of TCGA data focusing on HMTs found that the increased expression of G9a/EHMT2, an H3K9 methyltransferase, is associated with poor prognosis, while its oncogenic effect may be mediated by activating the expression of MTOR via increasing promoter H3K9me129,30,31.
PRC2, which catalyzes H3K27me3, is also overexpressed in GC, and this overexpression is associated with poor prognosis32. Recent studies have demonstrated that PRC2 may promote GC development via multiple pathways. Xing et al. found that EZH2 represses EBF1 expression by increasing promoter H3K27me, which subsequently activates the expression of TERT33, while another study showed that EZH2 may promote cancer via the downregulation of PTEN34. EZH2 was also found to induce H3K27me3 at the promoter of P21, a cell cycle regulator, and downregulate its expression35. In addition, EZH2 repressed the expression of ERRγ, a nuclear hormone receptor and a transcription factor, via H3K27me3, which led to the activation of an oncogene, FOXM1, in GC cells36.
Other HMTs have also been studied in recent years. For example, WDR5 is a subunit of the KMT2/MLL complex that catalyzes H3K4me1 and H3K4me2, and its abnormal expression is associated with poor prognosis. Mechanistically, it increased H3K4me3 at the promoter of CYCLIN D1, a cell cycle gene frequently upregulated in human cancer, and activated its expression37. KMT2 family genes were found to be mutated in approximately 10% of gastric cancer cases, and they were associated with PD-L1 expression, suggesting their potential roles in immune checkpoint therapy38. SETD1A, another HMT for H3K4, was overexpressed and localized to the promoter of SNAIL, a snail family transcriptional repressor, enhancing epithelial–mesenchymal transition (EMT) in GC cells39. In addition, the increased expression levels of DOT1L, an H3K79 methyltransferase, and protein arginine methyltransferases, such as PRM5 and PRMT6, were also associated with poor prognosis40,41,42. Together, these recent studies demonstrate that writer proteins regulate histone modification across the GC genome and that their increased expression likely leads to the dysregulation of multiple genes and signaling pathways, promoting cancer development.
Histone demethylases
LSD1, the first histone demethylase identified, has been shown to act on both H3K4 and H3K9, thus having the potential to both repress and activate target gene expression. LSD1 was found to be more highly expressed in GC than in normal tissue, and its increased expression was associated with metastasis and advanced cancer stages43,44. Mechanistically, it may promote EMT through the demethylation of promoter H3K4me2 and the subsequent downregulation of CDH1 (E-cadherin) expression44.
Histone deacetylases, acetyltransferases, and kinases
When the expression levels of histone deacetylases and kinases were analyzed, the upregulation of HDAC2, NEK6, and AURKA was identified in GC45,46. Corroborating these findings, a recent study showed that HDAC2 may mediate the deacetylation of H3K9 at the P21 promoter, leading to increased proliferation in GC cells47. In addition, the expression of the HATs P300 and CBP was found to be upregulated in GC cells48. Together, these recent studies have shown that altered expression levels of histone modifiers are associated with gastric tumorigenesis, leading to cancer progression through the regulation of oncogene and tumor suppressor gene expression.
Writers, erasers, and readers play a critical role in maintaining the proper chromatin state and gene expression for normal homeostasis. Mutations or altered expression levels of writers, erasers, and readers can lead to dysregulation of key signaling pathways and/or genes involved in gastric tumorigenesis (Fig. 1). The chromatin landscape, as defined by histone marks, is significantly remodeled during carcinogenesis. Therefore, new studies may benefit from genomic analyses of chromatin marks that are altered by the gain or loss of chromatin modifying factors. In addition, as most recent studies have utilized cancer cell lines, in vivo animal models or organs-on-a-chip combined with intact niche components would likely provide important, new insight into tumor heterogeneity and the microenvironment.
Writer and eraser proteins modify the histone tail by adding or removing modifications such as methylation (Me) or acetylation (Ac). These modifications are recognized by reader proteins, which are found in chromatin remodeling complexes capable of altering chromatin accessibility and allowing the binding of transcription factors to regulate gene expression. In GC, the expression of these proteins is dysregulated, leading to the up- and downregulation of oncogenes and tumor suppressors.
Alterations in chromatin remodeling complexes play functional roles in gastric carcinogenesis
One way in which the histone code can direct functional rearrangement of chromatin is via the recruitment of chromatin remodeling complexes. Currently, as defined by their core ATPase subunits, four subfamilies of chromatin remodeling complexes exist: SWI/SNF, INO80, CHD, and ISWI25. Of note, in each of these complexes, genetic alterations and changes in gene expression have been linked to GC development.
CHD
The chromodomain helicase DNA-binding (CHD) subfamily of chromatin remodeling factors is so named because of the presence of the chromodomains that interact with methylated histones. The expression of CHDs has been examined in human GC cases. Notably, mutations and reduced expression of CHD4 and CHD8 were found in microsatellite instability (MSI)-high GC49. Consistently, knockdown of CHD8 showed increased proliferation of GC cells50. The reduced expression of CHD5 also correlated with poor prognosis and invasion51. CHDs can interact with other epigenetic regulators, such as MBDs and MTAs, to form the NuRD complex, and subunits of this complex are involved in GC development. MBD2 was found to be downregulated in gastric preneoplastic lesions and tumors52. In contrast, the increased expression levels of MTA1 and MTA2 were associated with GC invasion and metastasis53,54,55. Indeed, the overexpression of MTA2 promoted colony formation in GC cells, potentially via the upregulation of IL-1156. Interestingly, increased expression of MTA3 was also found in GC compared to normal tissue. These studies highlight the importance of the subunit composition of the NuRD complex in gastric tumorigenesis57.
SWI/SNF
Unlike CHD, the SWI/SNF complexes contain subunits harboring bromodomains, which recognize acetylated histones. Specifically, the SWI/SNF localizes to H3K27ac-labeled active enhancers to facilitate gene expression58. This can be accomplished via an increase in the accessibility to transcription factors, as the SWI/SNF complex has the ability to mobilize and shift nucleosomes59,60. Multiple subunits of the SWI/SNF complex have been associated with gastric cancer development61,62,63,64. Most notably, unbiased genome and exome sequencing studies have identified ARID1A, a gene encoding the DNA interacting subunit of the SWI/SNF complex65, as the second most frequently mutated gene after TP53 in GC5,66,67. Several clinical studies have shown that ARID1A expression negatively correlates with increased invasion and metastasis and a worse prognosis, further supporting its role68,69,70. Cell culture studies using GC cell lines have shown that ARID1A loss promotes GC cell proliferation and migration71,72,73,74. Consistent with these data, we generated novel gastric tumor mouse models and demonstrated a tumor suppressor role of Arid1a in vivo75.
Unlike ARID1A, the increased expression of BRG1, the ATPase subunit of the SWI/SNF complex, appeared to correlate with gastric cancer development and metastasis76. Consistently, stabilization of BRG1 promoted EMT via the upregulation of SNAIL77,78. Interestingly, the expression of its mutually exclusive subunit, BRM, was lost during gastric carcinogenesis, highlighting the complex interactions between different subunits of the same chromatin remodeling complex.
In addition to its role as a chromatin remodeling complex, the loss of SWI/SNF subunits led to the global remodeling of the H3K27ac landscape, suggesting that it can regulate H3K27ac58,79,80. This function of the SWI/SNF complex may be facilitated by its association with histone acetyltransferases and HDACs81. Alternatively, the SWI/SNF complex may increase chromatin accessibility for other HATs to deposit H3K27ac. Corroborating these data, our H3K27ac analysis of mice with the heterozygous deletion of Arid1a showed that the loss of the H3K27ac+ enhancers was associated with the p53 and apoptosis pathway genes, followed by the corresponding reduction of their expression75. This study demonstrates that Arid1a is required for the maintenance of active enhancers in genes involved in the tumor suppressor pathway, providing new mechanistic insight into the epigenetic regulation of GC (Fig. 2).
The chromatin remodeling complex, SWI/SNF, is known to recognize histone acetylation via its reader subunit. The complex utilizes ATP-dependent helicase activity to slide nucleosomes and increase chromatin accessibility. This allows other histone acetyltransferases (HATs) to maintain h3K27ac, a histone modification associated with active gene expression. The loss of one copy of Arid1a in gastric tumors results in loss of H3K27ac at the enhancer and subsequent downregulation of p53 target genes followed by suppression of apoptosis, leading to tumor progression.
Given that the SWI/SNF and PRC2 complexes may regulate a common substrate such as H3K27, they may interact to control gene expression. Indeed, their antagonistic relationship has been demonstrated in multiple systems82,83,84. Furthermore, EZH2 inhibition has been shown to provide a specific vulnerability for ARID1A-mutated ovarian and gastric cancers, further supporting the interactions between writers and readers in cancer84,85.
ISWI and INO80
Much less is known about the ISWI and INO80 complexes in the context of GC. The ISWI subfamily contains a few complexes, including RSF and NURF complexes. The high expression levels of RSF-1 (part of the RSF complex) were associated with poor prognosis in GC86, and knockdown of the NURF complex subunits showed tumor suppressive effects in GC cells, further supporting their roles in GC87. While the role of the INO80 complex in gastric carcinogenesis is relatively unexplored, it contains YY1 as a subunit88, a transcription factor known to activate multiple oncogenic pathways in GC89,90,91,92,93,94. However, whether this activation is mediated by INO80 has yet to be elucidated.
These recent studies have shown that mutations in chromatin remodeling complexes and/or their altered expression levels promote gastric carcinogenesis by abnormal activation or inhibition of oncogenic and tumor-suppressive pathways. These chromatin complexes also interact with writers and readers to establish and maintain the oncogenic histone code.
The oncogenic chromatin landscape in gastric cancer
Since histone modification is closely tied to transcriptional regulation, it acts as a common mechanism for the dysregulation of gene expression during cancer development. The gain of active histone marks at the promoters/enhancers of oncogenes can lead to their increased expression, while the gain of repressive marks at the promoters/enhancers of tumor suppressors can lead to their reduced expression. These changes are often mediated by alterations in the expression or localization of writer proteins6. Here, we discuss recent studies providing new insight into how the oncogenic chromatin landscape influences GC through the regulation of promoters and enhancers.
With the advent of next-generation sequencings, such as chromatin immunoprecipitation sequencing (ChIP-seq), scientists have been able to investigate genome-wide changes in histone modification during gastric carcinogenesis. Using Nano-ChIP-seq for H3K4me1 and H3K4me3 (enhancer- and promoter-associated histone marks, respectively) and H3K27ac (active histone mark), Muratani et al. identified hundreds of H3K4me3+ promoters and H3K4me1+ enhancers whose activity differs between GC and normal tissue95. They defined the promoters that are gained in cancer, demonstrating that genes associated with these promoters are enriched in cancer-related gene sets and have prognostic values. This study suggested that the activation of promoters may be a critical driver of carcinogenesis. In addition, by integrating the chromatin profiles from ENCODE, the authors also found an enrichment of the PRC2 subunits EZH2 and SUZ12 in the promoters that are gained/lost in cancer, highlighting the role of PRC2 in maintaining the cancer chromatin landscape.
Muratani et al. also identified the presence of alternative promoters in GC95. These promoters, not located at their canonical transcriptional start sites (TSSs), drive the expression of noncanonical transcripts and protein isoforms, which could lead to the activation of oncogenic pathways. Another Nano-ChIP-seq study by the same group further expanded on the mechanisms of alternative promoters. They found that the use of alternative promoters in GC led to an overall decrease in the expression of peptides having a strong affinity for MHC class 1 molecule, thus leading to decreased antigen presentation and potential immune evasion96. Recently, Sundar et al. performed single-cell RNA-seq in gastric tumors and integrated the results of a previous study to stratify the tumors based on alternate promoter usage. They found that tumors with low alternative promoter usage contain more infiltrating T cells than tumors with high alternative promoter usage, supporting the role of alternative promoters in immune evasion97. The presence of these alternative promoters has subsequently been confirmed across a number of cancer types in a recent large-scale RNA-seq study, suggesting that they represent one of several new mechanisms underlying dysregulated gene expression during carcinogenesis98. A more recent study utilized long-read RNA-seq with multiple GC cell lines and showed that noncanonical transcripts from alternative promoters may also have different coding sequences and 3’ UTRs than canonical transcripts. Furthermore, the authors found that the use of alternative promoters for specific cancer genes such as ARID1A is associated with different survival outcomes and molecular subtypes in GC99, suggesting that alternative promoters can be utilized to alter the activity of key readers involved in gastric carcinogenesis (Fig. 3).
Alternative promoters recognized by H3K4me are frequently identified in GC. These promoters activate the expression of tumor-specific RNA and protein isoforms. These isoforms are highly expressed in tumors. In some cases, these tumor protein isoforms can have gain of function that can promote tumor invasion (e.g., MET and RASA3). Rarely, tumor isoforms are found to be downregulated in cancer compared to control, as in the case of ARID1A, specifically in GC of the CIN subtype. Furthermore, the expression of these tumor isoforms can result in a lower relative expression of canonical peptides with high affinity to MHC I, leading to decreased antigen presentation and immune evasion.
In addition to this promoter regulation, enhancers also play a key role in GC development. Ooi et al. analyzed superenhancers, which are large enhancer regions characterized by high levels of active chromatin marks and coactivator binding. They found that superenhancers gained in GC compared to normal tissue are enriched in genes related to important cancer hallmarks, including invasion, angiogenesis, and resistance to cell death100. Another recent study employed paired-end ChIP-seq of H3K27ac to identify chromosomal rearrangements involving enhancers in GC. The authors identified rearrangements leading to enhancer hijacking, in which an enhancer from one region of the genome relocates to activate a gene not normally under its regulation. This led to the activation of oncogenes such as CCNE1101. These studies showed that functional elements on chromatin, as defined by histone marks, can be remodeled to cause dysregulation of gene expression in GC.
The tumor microenvironment (TME) plays an important role in providing tumors with the factors necessary to facilitate their growth and progression. The chromatin landscape of TME in gastric cancer has recently been explored. Maeda et al. examined cancer-associated fibroblasts (CAFs) in GC by performing H3K27me3 ChIP-seq and found that H3K27me3 (a repressive mark) is lost in CAFs for genes associated with the gastric stem cell niche, such as WNT5A when compared to normal fibroblasts102. In addition, a recent study found that SAA1, a secreted protein involved in the migration of GC cells, is upregulated in CAFs compared to normal fibroblasts, which may be mediated by increased H3K27ac at its enhancers and promoter103. These studies provide new evidence that GC development can be promoted by epigenetic changes in the tumor microenvironment. Together, genome-wide chromatin analyses of both GC cells and the TME have provided new mechanistic insight into how oncogenic and tumor suppressor pathways are epigenetically regulated in gastric tumorigenesis.
Noncoding RNAs in gastric cancer
Over the last decade, noncoding RNAs (ncRNAs) have emerged as key players in the epigenetic regulation of carcinogenesis. ncRNAs are classified into short and long noncoding RNAs (lncRNAs), both of which regulate the expression of many key signaling pathways and regulator genes. One type of extensively studied short noncoding RNAs in GC are microRNAs (miRNAs), which are RNAs less than 30 nucleotides in length that silence complementary mRNA by either mRNA cleavage or translation repression104. In contrast, long noncoding RNAs (lncRNAs) are more than 200 nucleotides long, and they possess regulatory roles in facilitating miRNA–mRNA–protein crosstalk105. Here, we discuss recent discoveries addressing the roles of miRNAs and lncRNAs in gastric tumorigenesis.
miRNAs regulate the components of key oncogenic pathways in gastric cancer
While expression profiling experiments previously identified differentially expressed miRNAs between GC and normal gastric cells in vitro106, analysis of human GC and preneoplastic lesions identified distinct miRNAs that are up- or downregulated at different stages of the disease107. These studies suggested that miRNAs may play key roles during each step of gastric carcinogenesis.
By regulating key oncogenic pathways such as PI3K/AKT signaling, miRNAs can act as both tumor suppressors and oncogenes108,109,110. For example, miR137 acts as a tumor suppressor by repressing AKT, whereas miR-21, miR-221, and miR-222 promote gastric tumor cell proliferation by targeting PTEN, a negative regulator of AKT111,112. Additional miRNAs involved in the PI3K/AKT pathway have been identified. miR-107 promoted GC cell proliferation and metastasis by targeting FOXO1, a transcription factor downstream of AKT signaling, and FAT4, a tumor suppressor suppressing PI3K/AKT signaling113,114. Other miRNAs, such as miR-575 and miR-32-5p, were found to target PTEN, either directly or indirectly115,116. Additionally, by targeting the components of PI3K/AKT signaling, functional studies showed that miR-20b and miR-451a have opposing roles in gastric carcinogenesis117.
Another key oncogenic pathway regulated by miRNAs in GC is the Wnt signaling pathway, which is activated in approximately 46% of GC cases118,119. In a recent study, Deng et al. found significant upregulation of miR-192 and miR-215 in GC tissue samples and showed that these miRNAs activate Wnt signaling by targeting APC, a negative regulator of the pathway120. Another study illustrated the tumor suppressive role of miR-520f-3p in GC by targeting SOX9, an HMG-box transcription factor, and subsequently inactivating Wnt signaling121. These findings demonstrate that miRNAs regulate key oncogenic signaling pathways and that their expression is tightly regulated in gastric tumorigenesis.
LncRNAs regulate gastric tumorigenesis by facilitating crosstalk between miRNAs and their target genes
By interacting with DNA, mRNA, miRNA, and histone-modifying complexes, lncRNAs can regulate transcription, post-transcription, translation, and epigenetic modifications. They play key roles in tumorigenesis and have been shown to regulate cell proliferation, the cell cycle, apoptosis and metastasis122. Several lncRNA expression profile studies found aberrant expression patterns of lncRNAs in GC. A comparison of lncRNA expression profiles in gastric tumor tissues to adjacent nontumorous tissues identified two differentially expressed lncRNAs, uc001lsz and H19. As the second most downregulated lncRNA in gastric cancer, uc001lsz was proposed to have a tumor-suppressive, trans-acting effect on MUC2, which is highly expressed in GC. In contrast, the authors found that H19 was the most upregulated lncRNA in gastric tumor samples123. Consistent with this expression pattern, H19 is involved in multiple processes of gastric tumorigenesis. For example, Yang et al. demonstrated that H19 partially inactivates p53 in GC cells, while another study showed that the ectopic expression of H19 promotes gastric tumorigenesis by acting as a precursor of miR-675124,125. Other studies have also demonstrated that the H19/miR-675 axis promotes GC cell proliferation by targeting the tumor suppressor RUNX1126,127. In addition to encoding miR-675, H19 also interacts with other miRNAs to promote gastric tumorigenesis. For example, H19 promoted tumorigenesis via the miR-138/E2F2 axis128,129. Moreover, the upregulation of miR-22-3p rescued H19-induced GC cell growth130.
HOTAIR, another lncRNA upregulated in GC, was shown to promote tumorigenesis through its interactions with miRNAs and target proteins131,132,133. Specifically, HOTAIR promoted proliferation by negatively regulating miR-126 and activating CXCR4/RhoA signaling132 and enhanced gastric cancer cell invasion by inducing the ubiquitination of RUNX3, an upstream regulator of the tight junction protein CLAUDIN1133. Recently, HOTAIR has also been reported to promote gastric tumorigenesis by downregulating the exosomal levels of miR-30a and miR-30b, providing insight into the extracellular role of HOTAIR in GC134.
In addition to H19 and HOTAIR, other lncRNAs identified from lncRNA profiling in GC are beginning to gain attention135,136. For example, PVT1 has been shown to promote gastric tumorigenesis through its interactions with FOXM1, miR-125, and miR-30a137,138,139. Enriched in advanced GC140, MALAT1 has been shown to promote GC cell proliferation by recruiting SF2/ASF, a splicing factor, downregulating miR-204, and activating the PI3K/AKT pathway141,142,143,144. In summary, these studies show that the expression levels of specific lncRNAs are tightly regulated in GC tumorigenesis and that they interact with both miRNAs and target proteins to promote cancer progression.
Interaction between ncRNAs and histone modification in gastric cancer
The different modes of regulation in cells do not function in isolation but are interwoven in an intricate network to cooperatively regulate gene expression. As our understanding of both histone modification and ncRNAs grows, increasing interest is also focused on how their interactions control gastric tumorigenesis. Despite their distinct mechanisms of action, both miRNAs and lncRNAs influence histone modifications and are in turn regulated by these modifications in a reciprocal manner (Fig. 4).
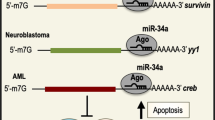
A miRNAs regulate the expression of histone modifiers, which are required to maintain the chromatin landscape. miRNAs themselves are regulated by histone modifiers. miRNA and histone modifiers can become dysregulated in GC, leading to the altered expression of oncogenes and tumor suppressors. B LncRNAs guide histone modifiers such as PRC2 and LSD1 to their target genes. Changes in lncRNA expression in GC lead to the dysregulated expression of their target oncogenes and tumor suppressors.
miRNAs and histone modifiers reciprocally regulate each other to promote or repress gastric tumorigenesis
Recent studies have examined the interactions between miRNAs and histone writers, demonstrating that they regulate each other in a reciprocal manner to promote or suppress tumorigenesis. miR-130b-3p promoted GC proliferation by inhibiting MLL3a, an H3K4 methyltransferase, in M2 macrophages145, while miR-4513 stimulated both GC proliferation and EMT by targeting KAT6B146.
In addition to histone writers, miRNAs also interact with erasers to either suppress or promote gastric tumorigenesis. For example, the regulation of KDM by miRNAs is well characterized for its tumor-suppressing effect. miR-194 was found to target KDM5B, and its overexpression suppressed GC growth both in vitro and in vivo147. Similarly, the upregulation of miR-29b suppressed gastric tumorigenesis by targeting KDM2A148. More recently, miR-329 and miR-491-5p were identified as regulators of KDM1A and KDM4B (JMJD2B) in suppressing tumorigenesis, respectively149,150. In contrast, miR-543 and miR-204 promoted proliferation and reversed SIRT1-induced EMT in GC cells by downregulating SIRT1, respectively151,152.
Alternatively, chromatin modifiers are also capable of regulating miRNAs in GC. For example, SIRT7-mediated H3K18ac repressed miR-34a, leading to the inhibition of apoptosis in GC cells153. A recent study also showed that BRD4 represses miR-216a-3p, a negative regulator of Wnt3a, thereby promoting the stemness of GC cells through the upregulation of Wnt signaling154. Additionally, deletion of HDAC1 led to an upregulation of miR-34a, resulting in the suppression of miR-34a’s downstream oncogene CD44, thereby inhibiting the metastasis of GC cells155. In contrast, deletion of LSD1 resulted in the downregulation of miR-142-5p, which led to the upregulation of its tumor-suppressing target CD9, thereby repressing gastric cancer cell migration43. These results highlight the existence of a complex miRNA-histone modifier network in which miRNAs and histone modifiers reciprocally regulate each other in gastric tumorigenesis.
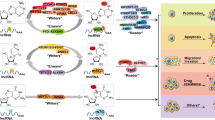
lncRNAs act as guides and/or scaffolds for histone modifiers
While miRNAs typically regulate the expression of histone-modifying proteins and complexes, lncRNAs interact directly/indirectly with writers and erasers, often recruiting them to their target genes to perform histone modifications. In particular, multiple lncRNAs interact with EZH2, a subunit of PRC2, to either attenuate or promote gastric tumorigenesis. A screen for lncRNAs in GC patients identified LINC00628 as a tumor suppressor, and LINC00628 suppressed tumorigenesis by interacting with EZH2 and guiding PCR2 to oncogenes such as CCNA2 and HOX11, leading to their methylation and silencing156. A more recent study suggested that lncRNAs can regulate gene expression by indirectly recruiting EZH2 through a mediator. In this study, Han et al. showed that lncRNA PART1, a lncRNA significantly downregulated in GC, upregulates PLZF via its interaction with androgen receptor (AR). PLZF then recruits EZH2 to increase H3K27 trimethylation at the PDGFB promoter, suppressing its expression and subsequently downregulating PI3K/Akt signaling157. Two additional lncRNAs, LINC01232 and LINC00202, were found to be upregulated in GC, and they interacted with EZH2, negatively regulating the expression of KLF2, a zinc-finger transcription factor, through histone methylation158,159. Another oncogenic lncRNA, HOTAIR, promoted EMT by recruiting PRC2 to catalyze H3K27me3 at the CDH1 promoter in GC160. Interestingly, EZH2 was recently shown to act upstream of lncRNAs. Zhu et al. demonstrated that EZH2 could methylate the promoter of a lncRNA, PCAT18, possibly contributing to its low expression in GC. The authors showed that PCAT18 could inhibit GC cell proliferation by regulating P16 expression161. These studies highlight the importance of ncRNA-histone writer interactions in gastric tumorigenesis.
Interactions between lncRNAs and erasers, specifically demethylases such as LSD1, also play crucial roles in gastric tumorigenesis. A recent study identified the upregulation of LINC00461 in GC tissues and showed that its knockdown in GC cells decreases cell viability and proliferation. Further evidence indicated that LINC00461 mediates GC cell viability via direct interaction with LSD1162. In light of this finding, more recent studies have uncovered the downstream mechanisms of lncRNA interactions with LSD1. For example, Lian et al. showed that LINC01446 promotes GC cell proliferation and metastasis by epigenetically silencing RASD1, a member of the Ras family of monomeric G proteins, through the recruitment of LSD1 to the RASD1 promoter163. In contrast, Pan et al. found that the binding of LINC00675 to LSD1 suppresses the transcription of SPRY4, an upstream effector of RAS, thereby reducing GC cell proliferation and migration. Interestingly, the overexpression of LINC00675 led to a stronger binding capacity of LSD1 to H3K4me2 and decreased H3K4me2 levels at the SPRY4 promoter, suggesting that LINC00675 regulates SPRY4 expression through LSD1 in GC164.
Interestingly, some lncRNAs can interact with both writer and eraser, highlighting their complex mechanistic roles in gastric tumorigenesis. For example, LINC00673 promoted GC cell proliferation by repressing key tumor suppressors, such as KLF2 and LATS2, through its interaction with EZH2 and LSD1. Specifically, LINC00673 facilitated EZH2 and LSD1 binding to the promoters of KLF2 and LATS2 to induce H3K27 trimethylation and H3K4 demethylation, respectively165. Another study found that FOXD2-AS1 promotes GC progression by recruiting both EZH2 and LSD1 to repress EPHB3 expression166. These results provide compelling evidence that lncRNAs can interact with multiple histone modifiers to regulate gastric tumorigenesis.
Recent studies have shown that lncRNAs are also able to act as scaffolds for multiple histone modifiers. For example, GClnc1 interacted with both WDR5 and KAT2A. GClnc1 promoted gastric tumorigenesis by acting as a scaffold for WDR5/KAT2A and recruiting them to the promoter of SOD2, a potential oncogene, which subsequently facilitated its expression by increasing H3K4me3167. Another study showed that GCAWKR recruits WDR5/KAT2A to upregulate the oncogene PTP4A1168. In addition to GClnc1 and GCAWKR, HOXA11-AS can also act as a scaffold for both histone modifiers and DNA methyltransferases, which include EZH2, LSD1, and DNMT1. Specifically, HOXA11-AS interacts with DNMT1/EZH2 to repress KLF2 expression through H3K27me3, while its interaction with LSD1/EZH2 represses PRSS8 expression through H3K27me3 and H3K4 demethylation169.
Finally, new studies have begun to explore lncRNA interactions with readers. For example, lncRNA NEAT1 directly interacted with BRG1, a subunit of the SWI/SNF complex, which resulted in the silencing of the G2/M checkpoint protein GADD45A and subsequently promoted GC cell proliferation170. In summary, these studies provide valuable insight into the diverse roles of lncRNA-histone modifier interactions, revealing a complex epigenetic network involved in the regulation of gastric tumorigenesis (Fig. 4).
Concluding remarks
The three main types of epigenetic regulation include DNA methylation, histone modification, and ncRNAs. While changes in DNA methylation remain one of the major mechanisms for gastric carcinogenesis, recent studies have shown that histone modifications and ncRNA also play critical roles. In this review, we discuss recent findings addressing the mechanism of histone modification and ncRNAs in GC. Specific writer and eraser proteins control histone modification and interact with readers such as chromatin remodeling complexes. Their dysregulation in GC leads to altered expression levels of downstream oncogenes and tumor suppressor genes. In addition to histone modification, ncRNAs regulate many key signaling pathways and genes involved in the development of GC. Importantly, these histone modifications and ncRNAs also interact with and regulate each other.
While somatic mutations of oncogenes and tumor suppressors are thought to be the major cause of cancer, accumulating evidence has increasingly supported the importance of epigenetic regulation in carcinogenesis. Since certain environmental factors are known to increase the risk of GC, epigenetic regulation would likely serve as a critical link between the environment and gene expression. Of note, the stomach is constantly exposed to environmental factors. Infection with two gastric pathogens, Epstein‒Barr virus (EBV) and H. pylori, is known to increase the risk of GC11,13. EBV in particular is known to cause widespread DNA methylation in GC, suggesting that repression of tumor suppressors by promoter hypermethylation is a core mechanism in EBV-mediated carcinogenesis15. Notably, a few recent studies have begun to explore the global changes in the chromatin landscape caused by EBV infection. Hi-C analysis revealed physical interactions between the EBV genome and the host genome, demonstrating EBV-mediated global changes in histone modification171. Similar to EBV-mediated DNA methylation, H. pylori can influence the expression of key regulators through altered DNA methylation172. It is also known to cause dysregulation of writers, readers, miRNAs, and ncRNAs, highlighting its potential in modifying the epigenetic landscape globally173,174,175,176,177,178,179,180. However, future studies are required to further define the mechanistic interactions between these environmental factors and epigenetic regulators.
New technologies continue to facilitate discoveries that will improve our understanding and treatment of GC. For example, combinatorial drug therapies prove to be a promising personalized medicine approach with the potential to reduce off-target toxicity181. In recent years, in vitro studies combining epigenetic inhibitors with other anticancer compounds have shown synergistic effects in tumor suppression for GC75,182,183,184. However, clinical trials investigating combinatorial treatment involving epigenetic inhibitors in GC have been limited185. In addition, novel single-cell technologies such as single-cell ATAC-seq allows us to probe the role of chromatin architecture and ncRNAs in tumor heterogeneity, which poses another major challenge in cancer treatment. Therefore, efforts to elucidate the epigenetic mechanism underlying gastric carcinogenesis will continue to improve the treatment and prevention of GC.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 71, 209–249 (2021).
Wong, M. C. S. et al. Global incidence and mortality of gastric cancer, 1980–2018. JAMA Netw. Open 4, e2118457 (2021).
Cervantes, A., Roda, D., Tarazona, N., Rosello, S. & Perez-Fidalgo, J. A. Current questions for the treatment of advanced gastric cancer. Cancer Treat. Rev. 39, 60–67 (2013).
Tan, P. & Yeoh, K. G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology 149, 1153–1162.e1153 (2015).
Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Padmanabhan, N., Ushijima, T. & Tan, P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat. Rev. Gastroenterol. Hepatol. 14, 467–478 (2017).
Yoda, Y. et al. Integrated analysis of cancer-related pathways affected by genetic and epigenetic alterations in gastric cancer. Gastric Cancer 18, 65–76 (2015).
Yamashita, S. et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc. Natl Acad. Sci. USA 115, 1328–1333 (2018).
Parsonnet, J. et al. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325, 1127–1131 (1991).
Nomura, A. et al. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 325, 1132–1136 (1991).
Uemura, N. et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345, 784–789 (2001).
Bae, J. M. & Kim, E. H. Epstein-Barr virus and gastric cancer risk: a meta-analysis with meta-regression of case-control studies. J. Prev. Med. Public Health 49, 97–107 (2016).
Tavakoli, A. et al. Association between Epstein-Barr virus infection and gastric cancer: a systematic review and meta-analysis. BMC Cancer 20, 493 (2020).
Li, Y., Liang, J. & Hou, P. Hypermethylation in gastric cancer. Clin. Chim. Acta 448, 124–132 (2015).
Matsusaka, K., Funata, S., Fukayama, M. & Kaneda, A. DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein-Barr virus. World J. Gastroenterol. 20, 3916–3926 (2014).
Fu, D. G. Epigenetic alterations in gastric cancer (Review). Mol. Med. Rep. 12, 3223–3230 (2015).
Kang, C., Song, J. J., Lee, J. & Kim, M. Y. Epigenetics: an emerging player in gastric cancer. World J. Gastroenterol. 20, 6433–6447 (2014).
Patel, T. N., Roy, S. & Ravi, R. Gastric cancer and related epigenetic alterations. Ecancermedicalscience 11, 714 (2017).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Hyun, K., Jeon, J., Park, K. & Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49, e324 (2017).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Hota, S. K. & Bruneau, B. G. ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882–2897 (2016).
Calcagno, D. Q., Gigek, C. O., Chen, E. S., Burbano, R. R. & Smith Mde, A. DNA and histone methylation in gastric carcinogenesis. World J. Gastroenterol. 19, 1182–1192 (2013).
Tie, J., Zhang, X. & Fan, D. Epigenetic roles in the malignant transformation of gastric mucosal cells. Cell. Mol. Life Sci. 73, 4599–4610 (2016).
Meng, X. et al. Comprehensive analysis of histone modification-associated genes on differential gene expression and prognosis in gastric cancer. Exp. Ther. Med. 18, 2219–2230 (2019).
Reyes, D. A., Sarria, V. M. S., Salazar-Viedma, M. & D’Afonseca, V. Histone methyltransferases useful in gastric cancer research. Cancer Inf. 20, 11769351211039862 (2021).
Chen, P. et al. Increased expression of EHMT2 associated with H3K9me2 level contributes to the poor prognosis of gastric cancer. Oncol. Lett. 20, 1734–1742 (2020).
Yin, C. et al. G9a promotes cell proliferation and suppresses autophagy in gastric cancer by directly activating mTOR. FASEB J. 33, 14036–14050 (2019).
He, L. J. et al. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac. J. Cancer Prev. 13, 3173–3178 (2012).
Xing, M. et al. Genomic and epigenomic EBF1 alterations modulate TERT expression in gastric cancer. J. Clin. Investig. 130, 3005–3020 (2020).
Gan, L. et al. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J. Hematol. Oncol. 11, 9 (2018).
Xu, J. et al. EZH2 promotes gastric cancer cells proliferation by repressing p21 expression. Pathol. Res. Pract. 215, 152374 (2019).
Huang, B. et al. Inhibition of EZH2 and activation of ERRgamma synergistically suppresses gastric cancer by inhibiting FOXM1 signaling pathway. Gastric Cancer 24, 72–84 (2021).
Sun, W., Guo, F. & Liu, M. Up-regulated WDR5 promotes gastric cancer formation by induced cyclin D1 expression. J. Cell. Biochem. 119, 3304–3316 (2018).
Wang, J. et al. Large-scale analysis of KMT2 mutations defines a distinctive molecular subset with treatment implication in gastric cancer. Oncogene 40, 4894–4905 (2021).
Wu, J. et al. Histone methyltransferase SETD1A induces epithelial-mesenchymal transition to promote invasion and metastasis through epigenetic reprogramming of snail in gastric cancer. Front. Cell Dev. Biol. 9, 657888 (2021).
Song, Z. et al. The role of DOT1L in the proliferation and prognosis of gastric cancer. Biosci. Rep. 40, BSR20193515 (2020).
Okuno, K. et al. Asymmetric dimethylation at histone H3 arginine 2 by PRMT6 in gastric cancer progression. Carcinogenesis 40, 15–26 (2019).
Liu, M. et al. PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics 10, 4437–4452 (2020).
Zhao, L. J. et al. LSD1 deletion represses gastric cancer migration by upregulating a novel miR-142-5p target protein CD9. Pharmacol. Res. 159, 104991 (2020).
Zhang, J. et al. Upregulation of LSD1 promotes migration and invasion in gastric cancer through facilitating EMT. Cancer Manag. Res. 11, 4481–4491 (2019).
Orenay-Boyacioglu, S. et al. Expression profiles of histone modification genes in gastric cancer progression. Mol. Biol. Rep. 45, 2275–2282 (2018).
Liu, X. et al. AURKA induces EMT by regulating histone modification through Wnt/beta-catenin and PI3K/Akt signaling pathway in gastric cancer. Oncotarget 7, 33152–33164 (2016).
Yu, Y. et al. RBBP8/CtIP suppresses P21 expression by interacting with CtBP and BRCA1 in gastric cancer. Oncogene 39, 1273–1289 (2020).
Wang, Y. M. et al. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 51, 1860–1868 (2017).
Kim, M. S., Chung, N. G., Kang, M. R., Yoo, N. J. & Lee, S. H. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58, 660–668 (2011).
Sawada, G. et al. CHD8 is an independent prognostic indicator that regulates Wnt/beta-catenin signaling and the cell cycle in gastric cancer. Oncol. Rep. 30, 1137–1142 (2013).
Hashimoto, T. et al. Clinical significance of chromatin remodeling factor CHD5 expression in gastric cancer. Oncol. Lett. 19, 1066–1073 (2020).
Pontes, T. B. et al. Reduced mRNA expression levels of MBD2 and MBD3 in gastric carcinogenesis. Tumour Biol. 35, 3447–3453 (2014).
Toh, Y. et al. Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int. J. Cancer 74, 459–463 (1997).
Lv, Z. Y. et al. Metastasis-associated protein 1 (MTA1) in gastric cancer tissues is positively associated with poorer prognosis. Pathol. Res. Pract. 214, 536–541 (2018).
Zhou, C. et al. MTA2 promotes gastric cancer cells invasion and is transcriptionally regulated by Sp1. Mol. Cancer 12, 102 (2013).
Zhou, C. et al. MTA2 enhances colony formation and tumor growth of gastric cancer cells through IL-11. BMC Cancer 15, 343 (2015).
Dong, H. et al. The metastasis-associated gene MTA3, a component of the Mi-2/NuRD transcriptional repression complex, predicts prognosis of gastroesophageal junction adenocarcinoma. PLoS ONE 8, e62986 (2013).
Wang, X. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Kassabov, S. R., Zhang, B., Persinger, J. & Bartholomew, B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 11, 391–403 (2003).
Dechassa, M. L. et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38, 590–602 (2010).
Tsuruta, S. et al. Solid-type poorly differentiated adenocarcinoma of the stomach: deficiency of mismatch repair and SWI/SNF complex. Cancer Sci. 111, 1008–1019 (2020).
Tessier-Cloutier, B. et al. Loss of switch/sucrose non-fermenting complex protein expression in undifferentiated gastrointestinal and pancreatic carcinomas. Histopathology 77, 46–54 (2020).
Gluckstein, M. I. et al. Comprehensive immunohistochemical study of the SWI/SNF complex expression status in gastric cancer reveals an adverse prognosis of SWI/SNF deficiency in genomically stable gastric carcinomas. Cancers (Basel) 13, 3894 (2021).
Liu, H., Zhao, Y. R., Chen, B., Ge, Z. & Huang, J. S. High expression of SMARCE1 predicts poor prognosis and promotes cell growth and metastasis in gastric cancer. Cancer Manag. Res. 11, 3493–3509 (2019).
Chandler, R. L. et al. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol. Cell. Biol. 33, 265–280 (2013).
Wang, K. et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43, 1219–1223 (2011).
Zang, Z. J. et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 44, 570–574 (2012).
Fabbrini, M. S., Zoppe, M., Bollini, R. & Vitale, A. 1-Deoxymannojirimycin inhibits Golgi-mediated processing of glycoprotein in Xenopus oocytes. FEBS Lett. 234, 489–492 (1988).
Wiegand, K. C. et al. ARID1A/BAF250a as a prognostic marker for gastric carcinoma: a study of 2 cohorts. Hum. Pathol. 45, 1258–1268 (2014).
Han, N., Kim, M. A., Lee, H. S. & Kim, W. H. Loss of ARID1A expression is related to gastric cancer progression, Epstein-Barr virus infection, and mismatch repair deficiency. Appl. Immunohistochem. Mol. Morphol. 24, 320–325 (2016).
Nagl, N. G. Jr. et al. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 65, 9236–9244 (2005).
Zhang, Q. et al. Chromatin remodeling gene AT-rich interactive domain-containing protein 1A suppresses gastric cancer cell proliferation by targeting PIK3CA and PDK1. Oncotarget 7, 46127–46141 (2016).
Dong, X. et al. Loss of ARID1A activates mTOR signaling and SOX9 in gastric adenocarcinoma-rationale for targeting ARID1A deficiency. Gut 71, 467–478 (2022).
Yan, H. B. et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis 35, 867–876 (2014).
Loe, A. K. H. et al. Uncovering the dosage-dependent roles of Arid1a in gastric tumorigenesis for combinatorial drug therapy. J. Exp. Med. 218, e20200219 (2021).
Sentani, K. et al. Increased expression but not genetic alteration of BRG1, a component of the SWI/SNF complex, is associated with the advanced stage of human gastric carcinomas. Pathobiology 69, 315–320 (2001).
Huang, L. Y. et al. SCF(FBW7)-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat. Commun. 9, 3569 (2018).
Ailiken, G. et al. Post-transcriptional regulation of BRG1 by FIRDeltaexon2 in gastric cancer. Oncogenesis 9, 26 (2020).
Mathur, R. et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 49, 296–302 (2017).
Alver, B. H. et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 (2017).
Nagl, N. G. Jr., Wang, X., Patsialou, A., Van Scoy, M. & Moran, E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26, 752–763 (2007).
Ho, L. et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903–913 (2011).
Wilson, B. G. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18, 316–328 (2010).
Bitler, B. G. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 21, 231–238 (2015).
Yamada, L. et al. Selective sensitivity of EZH2 inhibitors based on synthetic lethality in ARID1A-deficient gastric cancer. Gastric Cancer 24, 60–71 (2021).
Hu, B. S., Yu, H. F., Zhao, G. & Zha, T. Z. High RSF-1 expression correlates with poor prognosis in patients with gastric adenocarcinoma. Int. J. Clin. Exp. Pathol. 5, 668–673 (2012).
Ding, L. et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer 18, 45 (2019).
Cai, Y. et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874 (2007).
Wang, J. et al. The CCDC43–ADRM1 axis regulated by YY1, promotes proliferation and metastasis of gastric cancer. Cancer Lett. 482, 90–101 (2020).
Zhong, B. Z. et al. Effects of miR-384 and miR-134-5p acting on YY1 signaling transduction on biological function of gastric cancer cells. Onco Targets Ther. 13, 9631–9641 (2020).
Zheng, L. et al. miRNA-584-3p inhibits gastric cancer progression by repressing Yin Yang 1-facilitated MMP-14 expression. Sci. Rep. 7, 8967 (2017).
Bhaskar Rao, D., Panneerpandian, P., Balakrishnan, K. & Ganesan, K. YY1 regulated transcription-based stratification of gastric tumors and identification of potential therapeutic candidates. J. Cell Commun. Signal. 15, 251–267 (2021).
Kang, W. et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J. Transl. Med. 12, 80 (2014).
Zhang, T. et al. YY1-modulated long non-coding RNA SNHG12 promotes gastric cancer metastasis by activating the miR-218-5p/YWHAZ axis. Int. J. Biol. Sci. 17, 1629–1643 (2021).
Muratani, M. et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat. Commun. 5, 4361 (2014).
Qamra, A. et al. Epigenomic promoter alterations amplify gene isoform and immunogenic diversity in gastric adenocarcinoma. Cancer Discov. 7, 630–651 (2017).
Sundar, R. et al. Epigenetic promoter alterations in GI tumour immune-editing and resistance to immune checkpoint inhibition. Gut 71, 1277–1288 (2022).
Demircioglu, D. et al. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell 178, 1465–1477.e1417 (2019).
Huang, K. K. et al. Long-read transcriptome sequencing reveals abundant promoter diversity in distinct molecular subtypes of gastric cancer. Genome Biol. 22, 44 (2021).
Ooi, W. F. et al. Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity. Nat. Commun. 7, 12983 (2016).
Ooi, W. F. et al. Integrated paired-end enhancer profiling and whole-genome sequencing reveals recurrent CCNE1 and IGF2 enhancer hijacking in primary gastric adenocarcinoma. Gut 69, 1039–1052 (2020).
Maeda, M. et al. Cancer cell niche factors secreted from cancer-associated fibroblast by loss of H3K27me3. Gut 69, 243–251 (2020).
Yasukawa, Y. et al. SAA1 is upregulated in gastric cancer-associated fibroblasts possibly by its enhancer activation. Carcinogenesis 42, 180–189 (2021).
Macfarlane, L. A. & Murphy, P. R. MicroRNA: biogenesis, function and role in cancer. Curr. Genom. 11, 537–561 (2010).
Yang, G., Lu, X. & Yuan, L. LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta 1839, 1097–1109 (2014).
Zhang, X. et al. Integrated miRNA profiling and bioinformatics analyses reveal potential causative miRNAs in gastric adenocarcinoma. Oncotarget 6, 32878–32889 (2015).
Hwang, J. et al. MicroRNA expression profiles in gastric carcinogenesis. Sci. Rep. 8, 14393 (2018).
Zhang, H. et al. Integrated analysis of the miRNA, gene and pathway regulatory network in gastric cancer. Oncol. Rep. 35, 1135–1146 (2016).
Pereira, A. et al. miRNome reveals new insights into the molecular biology of field cancerization in gastric cancer. Front. Genet. 10, 592 (2019).
Hu, M., Zhu, S., Xiong, S., Xue, X. & Zhou, X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol. Rep. 41, 1439–1454 (2019).
Wu, L. et al. MicroRNA-137 contributes to dampened tumorigenesis in human gastric cancer by targeting AKT2. PLoS ONE 10, e0130124 (2015).
Chun-Zhi, Z. et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10, 367 (2010).
Li, F. et al. Upregulation of microRNA-107 induces proliferation in human gastric cancer cells by targeting the transcription factor FOXO1. FEBS Lett. 588, 538–544 (2014).
Wang, L., Li, K., Wang, C., Shi, X. & Yang, H. miR-107 regulates growth and metastasis of gastric cancer cells via activation of the PI3K-AKT signaling pathway by down-regulating FAT4. Cancer Med. 8, 5264–5273 (2019).
Wang, Y. N. et al. MicroRNA-575 regulates development of gastric cancer by targeting PTEN. Biomed. Pharmacother. 113, 108716 (2019).
Wang, Q. et al. microRNA-32-5p targets KLF2 to promote gastric cancer by activating PI3K/AKT signaling pathway. Am. J. Transl. Res. 11, 4895–4908 (2019).
Streleckiene, G. et al. miR-20b and miR-451a are involved in gastric carcinogenesis through the PI3K/AKT/mTOR signaling pathway: data from gastric cancer patients, cell lines and Ins-Gas mouse model. Int. J. Mol. Sci. 21, 877 (2020).
Ashrafizadeh, M., Rafiei, H., Mohammadinejad, R., Farkhondeh, T. & Samarghandian, S. Wnt-regulating microRNAs role in gastric cancer malignancy. Life Sci. 250, 117547 (2020).
Ooi, C. H. et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 5, e1000676 (2009).
Deng, S. et al. miRNA-192 and -215 activate Wnt/beta-catenin signaling pathway in gastric cancer via APC. J. Cell. Physiol. 235, 6218–6229 (2020).
Chen, J. Q. et al. MicroRNA-520f-3p inhibits proliferation of gastric cancer cells via targeting SOX9 and thereby inactivating Wnt signaling. Sci. Rep. 10, 6197 (2020).
Li, T., Mo, X., Fu, L., Xiao, B. & Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 7, 8601–8612 (2016).
Song, H. et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 11, 225 (2013).
Yang, F. et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 279, 3159–3165 (2012).
Li, H. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5, 2318–2329 (2014).
Zhuang, M., Gao, W., Xu, J., Wang, P. & Shu, Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 448, 315–322 (2014).
Liu, G., Xiang, T., Wu, Q. F. & Wang, W. X. Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol. Res. 23, 99–107 (2016).
Zhou, X. et al. The interaction between MiR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell. Physiol. Biochem. 36, 1440–1452 (2015).
Yu, J. et al. H19 rises in gastric cancer and exerts a tumor-promoting function via miR-138/E2F2 axis. Cancer Manag. Res. 12, 13033–13042 (2020).
Gan, L., Lv, L. & Liao, S. Long noncoding RNA H19 regulates cell growth and metastasis via the miR223p/Snail1 axis in gastric cancer. Int. J. Oncol. 54, 2157–2168 (2019).
Wei, Z. et al. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer 23, 1018–1032 (2020).
Xiao, J. et al. lncRNA HOTAIR promotes gastric cancer proliferation and metastasis via targeting miR-126 to active CXCR4 and RhoA signaling pathway. Cancer Med. 8, 6768–6779 (2019).
Xue, M. et al. HOTAIR induces the ubiquitination of Runx3 by interacting with Mex3b and enhances the invasion of gastric cancer cells. Gastric Cancer 21, 756–764 (2018).
Zhang, J. et al. HOTAIR contributes to the carcinogenesis of gastric cancer via modulating cellular and exosomal miRNAs level. Cell Death Dis. 11, 780 (2020).
Cao, W. J., Wu, H. L., He, B. S., Zhang, Y. S. & Zhang, Z. Y. Analysis of long non-coding RNA expression profiles in gastric cancer. World J. Gastroenterol. 19, 3658–3664 (2013).
Esfandi, F., Taheri, M., Kholghi Oskooei, V. & Ghafouri-Fard, S. Long noncoding RNAs expression in gastric cancer. J. Cell. Biochem. 120, 13802–13809 (2019).
Xu, M. D. et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin. Cancer Res. 23, 2071–2080 (2017).
Niu, J., Song, X. & Zhang, X. Regulation of lncRNA PVT1 on miR-125 in metastasis of gastric cancer cells. Oncol. Lett. 19, 1261–1266 (2020).
Wang, L. et al. lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. J. Cell Physiol. 236, 536–548 (2021).
Wang, Y. et al. Microarray expression profile analysis of long non-coding RNAs of advanced stage human gastric cardia adenocarcinoma. Mol. Genet. Genom. 289, 291–302 (2014).
Wang, J. et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed. Pharmacother. 68, 557–564 (2014).
Shao, G. et al. Long non-coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA-204 expression in gastric cancer. Oncol. Lett. 19, 805–812 (2020).
Zhu, K., Ren, Q. & Zhao, Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol. Lett. 17, 5335–5342 (2019).
Dai, Q., Zhang, T. & Li, C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag. Res. 12, 1929–1939 (2020).
Zhang, Y., Meng, W., Yue, P. & Li, X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 39, 134 (2020).
Ding, H., Shi, Y., Liu, X. & Qiu, A. MicroRNA-4513 promotes gastric cancer cell proliferation and epithelial–mesenchymal transition through targeting KAT6B. Hum. Gene Ther. Clin. Dev. 30, 142–148 (2019).
Bao, J., Zou, J. H., Li, C. Y. & Zheng, G. Q. miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. Eur. Rev. Med. Pharmacol. Sci. 20, 4487–4493 (2016).
Kong, Y. et al. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 381, 138–148 (2016).
Cai, L., Chen, Q., Fang, S., Lian, M. & Cai, M. MicroRNA-329 inhibits cell proliferation and tumor growth while facilitates apoptosis via negative regulation of KDM1A in gastric cancer. J. Cell. Biochem. 119, 3338–3351 (2018).
Zhang, J. et al. MiRNA-491-5p inhibits cell proliferation, invasion and migration via targeting JMJD2B and serves as a potential biomarker in gastric cancer. Am. J. Transl. Res. 10, 525–534 (2018).
Li, J., Dong, G., Wang, B., Gao, W. & Yang, Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem. Biophys. Res. Commun. 469, 15–21 (2016).
Zhang, L., Wang, X. & Chen, P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer 13, 290 (2013).
Zhang, S. et al. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci. Rep. 5, 9787 (2015).
Song, H. et al. BRD4 promotes the stemness of gastric cancer cells via attenuating miR-216a-3p-mediated inhibition of Wnt/beta-catenin signaling. Eur. J. Pharmacol. 852, 189–197 (2019).
Lin, L. et al. Depletion of histone deacetylase 1 inhibits metastatic abilities of gastric cancer cells by regulating the miR-34a/CD44 pathway. Oncol. Rep. 34, 663–672 (2015).
Zhang, Z. Z. et al. Long non-coding RNA LINC00628 functions as a gastric cancer suppressor via long-range modulating the expression of cell cycle related genes. Sci. Rep. 6, 27435 (2016).
Han, H. et al. Long noncoding RNA PART1 restrains aggressive gastric cancer through the epigenetic silencing of PDGFB via the PLZF-mediated recruitment of EZH2. Oncogene 39, 6513–6528 (2020).
Xu, Q. et al. LINC00202 attenuates the progression of gastric cancer via suppressing expression level of KLF2. J. BUON 26, 506–512 (2021).
Liu, J. et al. LINC01232 promotes gastric cancer proliferation through interacting with EZH2 to inhibit the transcription of KLF2. J. Microbiol. Biotechnol. 31, 1358–1365 (2021).
Song, Y. et al. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 54, 77–86 (2019).
Zhu, L. et al. EZH2-mediated epigenetic suppression of lncRNA PCAT18 predicts a poor prognosis and regulates the expression of p16 by interacting with miR-570a-3p in gastric cancer. J. Cancer 12, 7069–7078 (2021).
Shi, X. et al. Knockdown of LINC00461 inhibits cell proliferation and induces apoptosis in gastric cancer by targeting LSD1. Eur. Rev. Med. Pharmacol. Sci. 23, 10769–10775 (2019).
Lian, Y. et al. Long intergenic non-protein-coding RNA 01446 facilitates the proliferation and metastasis of gastric cancer cells through interacting with the histone lysine-specific demethylase LSD1. Cell Death Dis. 11, 522 (2020).
Pan, Y. et al. LINC00675 suppresses cell proliferation and migration via downregulating the H3K4me2 level at the SPRY4 promoter in gastric cancer. Mol. Ther. Nucleic Acids 22, 766–778 (2020).
Huang, M. et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 25, 1014–1026 (2017).
Xu, T. P. et al. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene 37, 5020–5036 (2018).
Sun, T. T. et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 6, 784–801 (2016).
Ma, M. et al. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol. Ther. 26, 2658–2668 (2018).
Sun, M. et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 76, 6299–6310 (2016).
Ma, P. et al. KLF5-modulated lncRNA NEAT1 contributes to tumorigenesis by acting as a scaffold for BRG1 to silence GADD45A in gastric cancer. Mol. Ther. Nucleic Acids 22, 382–395 (2020).
Okabe, A. et al. Cross-species chromatin interactions drive transcriptional rewiring in Epstein-Barr virus-positive gastric adenocarcinoma. Nat. Genet. 52, 919–930 (2020).
Muhammad, J. S., Eladl, M. A. & Khoder, G. Helicobacter pylori-induced DNA methylation as an epigenetic modulator of gastric cancer: recent outcomes and future direction. Pathogens 8, 23 (2019).
Hayashi, Y. et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut 62, 1536–1546 (2013).
Yang, F. et al. NF-kappaB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis. 9, 12 (2018).
Shen, J. et al. Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1 to promote Helicobacter pylori-induced gastric carcinogenesis. Cancer Res. 75, 754–765 (2015).
Chang, H. et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver 9, 188–196 (2015).
Lee, J. W. et al. Differential microRNA expression between gastric cancer tissue and non-cancerous gastric mucosa according to Helicobacter pylori status. J. Cancer Prev. 22, 33–39 (2017).
Matsushima, K. et al. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer 128, 361–370 (2011).
Zhu, H. et al. Microarray analysis of Long non-coding RNA expression profiles in human gastric cells and tissues with Helicobacter pylori infection. BMC Med. Genom. 8, 84 (2015).
Chu, A., Liu, J., Yuan, Y. & Gong, Y. Comprehensive analysis of aberrantly expressed ceRNA network in gastric cancer with and without H. pylori infection. J. Cancer 10, 853–863 (2019).
Al-Lazikani, B., Banerji, U. & Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 30, 679–692 (2012).
Liu, S. et al. Diosgenin and GSK126 produce synergistic effects on epithelial–mesenchymal transition in gastric cancer cells by mediating EZH2 via the Rho/ROCK signaling pathway. Onco Targets Ther. 13, 5057–5067 (2020).
Regel, I. et al. Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology 143, 99–109.e110 (2012).
Park, J. K., Seo, J. S., Lee, S. K., Chan, K. K. & Kuh, H. J. Combinatorial antitumor activity of oxaliplatin with epigenetic modifying agents, 5-Aza-CdR and FK228, in human gastric cancer cells. Biomol. Ther. 26, 591–598 (2018).
Yoo, C. et al. Vorinostat in combination with capecitabine plus cisplatin as a first-line chemotherapy for patients with metastatic or unresectable gastric cancer: phase II study and biomarker analysis. Br. J. Cancer 114, 1185–1190 (2016).
Acknowledgements
We would like to thank Dr. Roshane Francis and Edward Espinoza for their feedback on our manuscript. This work has been supported by the Canadian Institutes of Health Research (CIHR PJT 173266) (T.-H.K.); SickKids Restracomp, Ontario Graduate and Canada Graduate Scholarships (A.K.H.L.).
Author information
Authors and Affiliations
Contributions
A.K.H.L. and L.Z. drafted the manuscript, and T.-H.K. edited the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Loe, A.K.H., Zhu, L. & Kim, TH. Chromatin and noncoding RNA-mediated mechanisms of gastric tumorigenesis. Exp Mol Med 55, 22–31 (2023). https://doi.org/10.1038/s12276-023-00926-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-023-00926-0
This article is cited by
-
Regulatory RNA: from molecular insights to therapeutic frontiers
Experimental & Molecular Medicine (2024)
-
Decoding the regulatory landscape of lncRNAs as potential diagnostic and prognostic biomarkers for gastric and colorectal cancers
Clinical and Experimental Medicine (2024)
-
Unveiling the genetic and epigenetic landscape of colorectal cancer: new insights into pathogenic pathways
Medical Oncology (2023)