Abstract
Cancer is one of the most difficult diseases in human society. Therefore, it is urgent for us to understand its pathogenesis and improve the cure rate. Exosomes are nanoscale membrane vesicles formed by a variety of cells through endocytosis. As a new means of intercellular information exchange, exosomes have attracted much attention. Noncoding RNAs exist in various cell compartments and participate in a variety of cellular reactions; in particular, they can be detected in exosomes bound to lipoproteins and free circulating molecules. Increasing evidence has suggested the potential roles of exosomal noncoding RNAs in the progression of tumors. Herein, we present a comprehensive update on the biological functions of exosomal noncoding RNAs in the development of cancer. Specifically, we mainly focus on the effects of exosomal noncoding RNAs, including microRNAs, circular RNAs, long noncoding RNAs, small nuclear RNAs, and small nucleolar RNAs, on tumor growth, metastasis, angiogenesis, and chemoresistance. Moreover, we outline the current clinical implications concerning exosomal noncoding RNAs in cancer treatment.
Similar content being viewed by others
Introduction
Since the twenty-first century, the incidence rate of cancer has increased at an alarming rate along with the changing lifestyles and habits and increased life expectancy of people. Current thinking suggests that cancer is a complicated disease caused by accumulating genomic and epigenetic aberrations1. There are eight biological capabilities acquired during the multistep development of human tumors to rationalize the complexities of neoplastic disease2. These processes are involved in sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, reprogramming energy metabolism, and evading immune destruction. Therefore, it is imperative to better understand the mechanisms of tumorigenesis and development to improve therapeutic efficacy.
Extracellular vesicles (EVs) are structures released by all types of cells into the environment3. They are enveloped by a phospholipid bimolecular layer and can be composed of a variety of components. Exosomes, a subtype of EVs, can deliver proteins, nucleic acids and noncoding RNAs (ncRNAs) to recipient cells for intercellular communication. Since certain proteins and nucleic acids can be transferred to receptor cells in the tumor microenvironment (TME), exosomes can function to transform healthy cells into cells with the characteristics of cancer cells and create a fertile environment for tumor cells to grow4. Currently, studies on the impact of exosomes on tumorigenesis and carcinoma progression have expanded5,6,7. In addition, due to the stability of exosomes and their low immunogenicity, exosomes have been developed as vehicles to carry drugs and antitumor nucleic acids for tumor therapy.
Approximately 50% of the DNA in the genome can be transcribed into RNA, of which only 2% can be translated into proteins, and the remaining 98%, known as ncRNAs, cannot be translated, including small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), circular RNAs (circRNAs), microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), long noncoding RNAs (lncRNAs) and small interfering RNAs (siRNAs). Through interaction with DNA, RNA, and proteins, ncRNAs can modulate the expression levels of genes at the transcriptional, posttranscriptional, and epigenetic levels, thus determining the fate of cells8,9. In this review, we mainly provide an overview of the roles of exosomal ncRNAs, especially exosomal miRNAs, exosomal circRNAs, and exosomal lncRNAs, in tumor progression, including tumor growth, metastasis, angiogenesis, and chemoresistance. At the same time, we briefly summarize the current application of exosomal ncRNAs in tumor therapy.
The biological characteristics of exosomes
The structure and biogenesis of exosomes
When the cell is activated or damaged, some of the proteins, nucleic acids, lipids, and other substances will be separated into multivesicular bodies (MVBs) and released to the extracellular matrix. According to the diameter, MVBs can be divided into three types10: (1) exosomes with a diameter of 30–150 nm; (2) microparticles with a diameter of 100–1000 nm; and (3) apoptotic bodies with a diameter of 1–4 μm11. Initially, exosomes were described by Trams et al. as exfoliated vesicles12, and then Harding and Stahl discovered them in rat reticulocytes13. Finally, Johnstone named these structures “exosomes” in 198714. Due to the lack of research methods, exosomes were once considered “garbage bags” produced by host cells to maintain the balance of cell components and secrete intracellular waste products without any biological functions14. Upon in-depth study of exosomes, however, it has been confirmed that mast cells, dendritic cells, lymphocytes, fibroblasts, mesenchymal stem cells, and tumor cells can all produce and release exosomes. A growing body of evidence has revealed that these “unnecessary compounds” actually have an important impact on physiological and pathological processes, such as damage repair15, antigen presentation16, tumor metastasis, and other important pathways17.
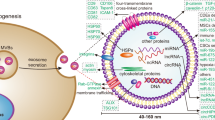
Exosomes are secreted in different ways from the other two MVBs18, as shown in Fig. 1. Both microparticles and apoptotic bodies are produced through the extension of the plasma membrane, while exosomes are produced in the form of intraluminal vesicles (ILVs). In the early stage of exosome formation, the plasma membrane buds inward and forms ILVs, also known as early endosomes. The endosomal sorting complex required for transport (ESCRT) protein family is responsible for recruiting cluster-specific substances and transporting them to the endosomal membrane. Four subtypes of ESCRT proteins, ESCRT0-III, work together to form late endosomes19. With the accumulation of cargoes and inward budding of endosome membranes, multiple ILVs containing proteins, RNA, and other substances are formed. Presently, they are called MVBs. MVBs can be used as transporters to deliver cargoes to lysosomes for degradation, or they can fuse with the plasma membrane through the regulation of Rab proteins to release ILVs, which are exosomes. Of course, there are various other regulators involved in the complex process of exosome formation20.
Exosome biogenesis begins with the inward budding of the plasma membrane and then the formation of early endosomes. With the assistance of the ESCRT protein family, a variety of DNA, RNA, and proteins gather to form late endosomes, further developing MVBs. Finally, MVBs are released into the extracellular environment by fusing with the plasma membrane to form exosomes. Both microparticles and apoptotic bodies arise through the extension of the plasma membrane.
The composition of exosomes
Biological methods such as differential centrifugation, sucrose gradient ultracentrifugation, transmission electron microscopy, western blotting, and mass spectrometry are used to analyze the composition and function of exosomes. Their contents include proteins, DNA, RNA, and even genetic materials of viruses and prions. Among them, proteins can be divided into specific proteins and nonspecific proteins. Most nonspecific proteins are derived from cytoplasmic and membrane proteins in the conserved regions of parental cells, which mainly maintain the structure and function of exosomes. Specific proteins are related to the origin of parent cells, such as exosomes from T lymphocytes with granular enzymes and perforin proteins on the surface21.
In addition to proteins, exosomes contain a large number of RNAs that can perform biological functions22. NcRNAs are biological molecules that cannot encode proteins (i.e., they have no protein-coding function) but can control gene expression and cell differentiation at the genomic and chromosomal levels. In recent decades, researchers have realized that ncRNAs are no longer “junk” transcriptional products but are biological molecules with regulatory functions that mediate various biological processes. Many ncRNAs can be recruited by ESCRT proteins to form exosomal ncRNAs. Through deep sequencing analysis, the ncRNAs identified from exosomes in human plasma and urine included snRNAs, snoRNAs, circRNAs, miRNAs, piRNAs, lncRNAs, ribosomal RNAs (rRNAs), and transfer RNAs23,24. In comparison with free ncRNAs, exosomal ncRNAs, as a new vector, can not only protect ncRNAs from enzyme degradation but also deliver ncRNAs to produce targeted biological effects25. We do not fully understand how exosomal ncRNAs exert their function in different tumors thus far, but there is no doubt that they play a crucial role in tumor progression. Exosomes are a double-edged sword, and in-depth research will pave the way for accurate and effective treatments.
The roles of exosomal miRNAs in cancer
Background on miRNAs
MiRNAs refer to single-stranded RNA molecules approximately 22 nucleotides in length that mainly regulate gene expression at the posttranscriptional level. MiRNAs can not only reduce the protein produced by transcripts but also reduce the level of the transcripts26. A single miRNA can target hundreds of mRNAs and influence the expression of target genes that are often involved in functional interacting pathways. In fact, many diseases are associated with unique miRNA characteristics, which indicates that specific miRNAs are activated in some pathophysiological processes27. The TME, where exosomes are released, is critical for the malignant phenotypes of tumors. Valadi et al. showed early on that exosomal miRNAs could be delivered to recipient cells to program relevant functions5. Moreover, Umezu et al. found that miRNAs mediated communication between neoplasia and endothelial cells28. Similarly, it has also been demonstrated that miRNAs are transferred from monocytes to endothelial cells and modulate endothelial cell function29. In the last decade, multiple studies have revealed that cancer-derived exosomal miRNAs play important roles in the recruitment and reprogramming of constituents of the tumor environment30. Since miRNAs are important gene expression regulators and candidate genes for the development of biomarkers, research on miRNAs is rapidly expanding.
Exosomal miRNAs in tumor growth
Tumor growth is a malignant phenotype of tumors; thus, increasing attention has been given to understanding the mechanisms of cell proliferation. Advances in miRNA research provide new opportunities for cancer treatment31,32. Our previous studies have shown that higher miR-377 expression is related to the progression of esophageal squamous cell carcinoma (ESCC), suggesting a novel direction for the treatment and diagnosis of ESCC33. Increasing studies have recently suggested a relationship between exosomal miRNAs and tumor growth. For example, a recent study on esophageal adenocarcinoma demonstrated that decreased levels of miR-30a-3p and miR-30a-5p were associated with the activation of the Wnt signaling pathway, enhancing cell proliferation and tumor growth34. In another study of hepatocellular carcinoma (HCC), cancer cells were shown to secrete nanovesicles and exosomes, which were different from their source cells35. In addition, exosomal miR-126 from endothelial cells was found to be related to the progression of non-small-cell lung cancer (NSCLC) cells and malignant mesothelioma36,37. Similarly, in ovarian cancer (OC), exosomal miRNA-454 was suggested to disrupt the Wnt pathway by targeting proline-rich transmembrane protein 2, thereby promoting OC cell growth in vivo38. Intriguingly, Melo et al. reported that breast cancer (BC)-derived exosomes contain miRNAs associated with the RISC-Loading Complex, which has the cell-independent capacity to process precursor miRNAs into mature miRNAs, thus promoting tumorigenesis39.
Exosomal miRNAs in tumor metastasis
Metastasis is an overwhelming cause of mortality in cancer patients. Unsurprisingly, exosomal miRNAs are also involved in tumor metastasis. Cancer-secreted miRNAs are emerging mediators of cancer-host crosstalk. For example, highly metastatic HCC cell-secreted exosomal miR-1247-3p directly targets B4GALT3 to activate β1-integrin-NF-κB signaling in fibroblasts, thus increasing the ability of normal fibroblasts to transform into CAFs40. In addition, exosomal miRNA-105, which is characteristically expressed and secreted by metastatic BC, has been shown to promote the metastasis of BC by targeting the tight junction protein ZO-141. Similarly, high miR-122 levels in the circulation have been proven to be associated with metastasis in BC42. In addition, miR-19a and miR-19b have been reported to be associated with the progression of liver metastasis in human colorectal cancer (CRC)43. One study suggested that the expression of 11 exosomal miRNAs in patients with metastatic PC changed significantly compared with that in patients with nonmetastatic PC, of which miR-375 and miR-200b were confirmed to be related to metastasis44. Another study on PC also showed a significant increase in exosomal miRNA-141 levels in patients with metastatic features45.
Exosomal miRNAs in tumor angiogenesis
Angiogenesis, the development of new blood vessels from preexisting vessels, plays an important role in tumor growth, maintenance and metastasis46. Increasing evidence suggests the potential role of exosomal miRNAs in tumor angiogenesis. For example, exosomal miR-9 has been found to effectively reduce SOCS5 levels and activate the JAK-STAT pathway, further promoting endothelial cell migration and tumor angiogenesis47. MiR-210 derived from exosomes was proven to be enriched in exosomes targeted to stromal cells. Upon further study, Cui et al. found that exosomal miR-210 reduces the expression of the miR-210 target protein Ephrin A3, thereby promoting lung adenocarcinoma angiogenesis48. Additionally, another study on exosomal miR-210 mediated by neutral sphingomyelinase 2 (nSMase2) suggested that it can be transported to endothelial cells and participate in the suppression of target genes, thereby enhancing angiogenic activities49. Moreover, a study showed that exosomal miR-21 leads to the activation of STAT3, which elevates the level of VEGF in human bronchial epithelial cells and is involved in the angiogenesis of human umbilical vein endothelial cells (HUVECs)50. Similarly, miR-221-3p derived from exosomes was found to be significantly enriched in cervical squamous cell carcinoma51.
Exosomal miRNAs in tumor chemoresistance
Chemoresistance challenges the clinical outcomes of cancer patients and remains the main obstacle to cancer therapy52. Recent studies have suggested that some miRNAs are likely to be responsible for drug resistance in various cancers. Camptothecin is a drug used in cancer chemotherapy to induce apoptosis. However, analyses have shown that overexpression of exosomal miR-373 inhibits the camptothecin-induced apoptosis of tumor cells53. In addition, the chemotherapy drug cisplatin has long been one of the first-line drugs for fighting tumors, especially those of the lung, ovary, and testes. Challagundla et al. found that exosomal miR-155 transferred by human monocytes can immediately target telomeric repeat binding factor 1 (TERF1) and elevate neuroblastoma cell resistance to cisplatin54. Likewise, 5-fluorouracil (5-FU) is also a crucial agent for adjuvant therapy after curative surgery55,56,57. A study showed that in the serum exosomes of drug-resistant CRC patients, the expression levels of some miRNAs, such as miR-21-5p and miR-1246, were significantly higher than those in the serum exosomes of sensitive control patients. The authors pointed out that targeting these miRNAs could promote the chemotherapy sensitivity of CRC cells, which may provide a new direction for CRC therapy58. However, miR-29c was shown to be frequently downregulated in the serum samples of patients with ESCC, which has also been shown to be related to the resistance of ESCC to 5-FU59.
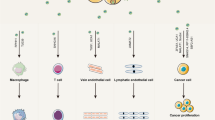
In general, the mechanism by which exosomal miRNAs promote tumor progression can be divided into the following categories: (a) Regulating the expression of genes involved in angiogenesis, such as VEGF60,61 (Fig. 2a); (b) Direct binding to the 3′ UTR of the target gene, thus affecting its function62,63 (Fig. 2b); (c) Interfering with the immune system and disrupting its function64,65 (Fig. 2c); (d) Participating in the EMT process66,67 (Fig. 2d); (e) Reprogramming metabolic balance68,69 (Fig. 2e); (f) Activating classical signaling pathways related to tumorigenesis and development70,71 (Fig. 2f). The corresponding exosomal miRNAs in each mechanism are shown as representatives in Fig. 2(a–f).
a Promoter angiogenesis. b Interacting with the 3′ UTR of its target genes and disrupting its transcription level. c Regulating the immune system as immunomodulators. d Influencing the progress of EMT. e Participating in tumor metabolic reprogramming. f Affecting the classic signaling pathways involved in tumorigenesis and development.
The roles of exosomal circRNAs in cancer
Background on circRNAs
CircRNAs are a novel class of ncRNA molecules that have recently become a research hotspot in the field of RNA. They are endogenous RNAs formed by alternative splicing and are widely distributed in eukaryotic cells. The structure of circRNA molecules is relatively stable and is different from that of linear RNA molecules because it is a closed circular molecule without 5′ and 3′ ends and is not affected by exonucleases72. Previous studies have proposed that circRNAs can serve as effective miRNA sponges, as they contain conserved miRNA targets, competitively inhibiting miRNA regulation of downstream target gene expression73,74,75. Moreover, studies have confirmed that circRNAs can be enriched in exosomes11,76. These molecules have various functions in the body and participate in many physiological processes, such as the proliferation and invasion of tumor cells and the progression of other diseases76.
Exosomal circRNAs in tumor growth
It has been reported that exosomal circRNAs function mainly by sponging miRNAs. A study from Yongchao Li et al. demonstrated that the circRNA FMN2 was highly abundant in the serum of CRC patients. Functional experiments and bioinformatics analysis indicated that the circRNA FMN2 promotes CRC proliferation via the miR-1182/hTERT signaling pathway77. In another study, the circRNA IFT80 was found to be significantly increased in the serum and cells of CRC patients. Silencing IFT80 inhibited CRC cell proliferation in vitro and in vivo. Mechanistically, the exosomal circRNA IFT80 exerts its function through the miR-1236-3p/HOXB7 axis78. In addition, Zhang et al. found that the adipose-secreted circRNA circ-DB regulates deubiquitination in HCC by controlling miR-34a and deubiquitination-related USP7, thereby promoting cell growth79. Similarly, a study using a xenograft mouse model of liver cancer cells revealed that tumor-derived exosomal circRNAs in serum were related to tumor mass. Exosomal circRNAs can maintain biological activity and abolish miRNA-induced growth inhibition in recipient cells80. In gastric cancer (GC), one study proved that knockdown of the circRNA NRIP1 can suppress the growth of GC cells. In terms of the mechanism, NRIP1 may act as a suppressor by sponging miR-149-5p, but this speculation needs further demonstration81. In another article, Xie et al. demonstrated that exosomal circSHKBP1 regulates the miR-582-3p/HUR/VEGF pathway, suppresses HSP90 degradation, and promotes GC progression82.
Exosomal circRNAs in tumor metastasis
Tumor metastasis requires not only growth and invasion abilities but also the ability of cancer cells to cross the endothelial barrier. The process of EMT is also regulated by many circRNAs, which indicates that circRNAs may have an important role in EMT-related processes, including metastasis83. At present, our understanding of the function of exosomal circRNAs in tumor metastasis and their molecular mechanisms is limited. A study pointed out that circ-RanGAP1 was significantly elevated in the tissues and plasma exosomes of patients with GC, which was related to the late stage of tumor node metastasis. A mechanistic study showed that circ-RanGAP1 elevated the expression levels of VEGF-A (VEGFA) by sponging miR-877-3p84. Similarly, the expression of exosomal circ-FECR was increased in small cell lung cancer (SCLC), which was associated with its metastasis85. Moreover, an analysis of the association between the levels of the circRNA ciRS-7 and the clinicopathological characteristics of HCC patients indicated that the levels of ciRS-7 were highly associated with hepatic microvascular invasion86. Intriguingly, circRNAs could be induced by hypoxia, which is one of the important properties of solid tumors. One article suggested that hypoxia-derived exosomal circ-133 was transported into normoxic cancer cells and promoted cell migration via the miR-133a/GEF-H1/RhoA axis87.
Exosomal circRNAs in tumor angiogenesis
CircRNAs have recently been regarded as a vital factor in cancer progression; however, the function of exosomal circRNAs in tumor angiogenesis has not been explored clearly. Specifically, few studies have examined the role of exosomal circRNAs in tumor angiogenesis, resulting in a largely unknown understanding of their underlying mechanisms, which urgently requires further exploration. Currently, only one study has shown that coculture of HUVECs and exosomes derived from HCC cell lines significantly enhances HUVEC angiogenesis and tumor metastasis. Further study found that exosomal circRNA-100,338, which was highly expressed in both highly metastatic HCC cells and their secreted exosomes, mediates angiogenesis88.
Exosomal circRNAs in tumor chemoresistance
As various potential mechanisms are involved in the progression of drug resistance, exosomes have attracted extensive attention as a new therapeutic approach. Recently, studies have indicated that circRNAs can regulate chemotherapy resistance in many kinds of cancers89. In SCLC, exosomal circRNA-FECR1 is considered to be important for chemoresistance. Data revealed that exosomal circRNA-FECR1 was highly abundant in the serum of SCLC patients compared with that of healthy controls and was clinically associated with poor survival and chemotherapy response85. Moreover, in glioma, one study revealed that the expression of exosomal circRNA NFIX was significantly increased in the serum of patients with temozolomide resistance. The exosomal circRNA NFIX mediates the resistance of gliomas to temozolomide at least in part by sponging miR-13290. Regarding CRC, a study suggested that PKM2 expression levels in CRC cells are heterogeneous; that is, PKM2 is abundant in oxaliplatin-resistant cells and relatively low in drug-sensitive cells. A mechanistic study indicated that once transferred from resistant cells to sensitive cells, the circRNA hsa-circ-0005963 could transmit oxaliplatin resistance by sponging miR-122 and upregulating PKM291. Furthermore, a microarray analysis of exosomal RNAs from chemoresistant and chemosensitive CRC cells was performed by Hon et al. to identify key exosomal RNAs associated with chemoresistant CRC. The results showed that exosomal circ_0000338 may have dual regulatory roles in chemoresistant CRC92. Recently, another study revealed that HCC-derived exosomal circUHRF1 contributes to immunosuppression by inducing NK cell dysfunction, thus driving resistance to anti-PD1 immunosuppression93.
Overall, abnormal expression of exosomal circRNAs has been found in various types of cancer. By gain- and loss-of-function assays, researchers have identified the role of exosomal circRNAs in the progression of cancer, which provides a better understanding of the relationship between exosomal circRNAs and cancer94,95,96,97,98,99,100,101,102,103 (Table 1). However, the mechanism of exosomal circRNAs in cancer deserves further study.
The roles of lncRNAs in cancer
Background on lncRNAs
LncRNAs generally refer to RNA transcripts longer than 200 nucleotides (Table 2). It was originally believed that lncRNAs were transcriptional byproducts of RNA polymerase II that could not be translated into proteins and had no biological functions. However, increasing amounts of evidence have proven that they actually participate in many biological reactions, such as chromatin remodeling, histone modification and X chromosome inactivation. In fact, by directly or indirectly interfering with gene expression in a variety of cancers, lncRNAs were demonstrated to play important roles in multiple biological processes, including cell growth, differentiation, and proliferation104. Notably, lncRNAs can bind to various biological macromolecules in cells, such as DNA, RNA, and proteins105. Similarly, via exosomes, lncRNAs can be transported into body fluids. Interestingly, some lncRNAs are enriched in exosomes, while others are virtually absent, suggesting that some lncRNAs are selectively classified into exosomes. Exosomal lncRNAs account for approximately 3% of the total RNA of exosomal species.
Exosomal lncRNAs in tumor growth
Many exosomal lncRNAs are involved in tumorigenesis and tumor growth. Some of them are downregulated in pathological samples to promote tumor growth. A study by Zhang et al. detected exosomal lncRNA-H19 content in the serum of healthy subjects and patients with pituitary tumors and found that the expression of lncRNA-H19 was significantly reduced in tumor patients. Further studies demonstrated that exosomal lncRNA-H19 could suppress distal tumor cell growth by inhibiting 4E-BP1 phosphorylation106. Additionally, the lncRNA SENP3-EIF4A1 was found to be significantly decreased in the serum of HCC patients. Research has shown that SENP3-EIF4A1 derived from exosomes could inhibit tumor growth in vivo and modulate the levels of ZFP36 ring finger protein (ZFP36) by competitively binding to miR-9-5p84. There are, of course, some lncRNAs that are abnormally upregulated in pathological samples. For example, the expression of the lncRNA prostate cancer-expressed EZH2-associated transcript (PCSEAT) was suggested to be specifically upregulated in prostate cancer (PCa)107. Similarly, the lncRNA BCRT1, a lncRNA that can remarkably suppress cancer progression both in vivo and in vitro, was markedly increased in BC tissues. In terms of the mechanism, lncRNA BCRT1 was able to competitively bind with miR-1303 and inhibit the degradation of the target gene polypyrimidine tract-binding protein 3 (PTBP3), which can promote the progression of BC108. These clues all suggest that exosomal lncRNAs could regulate cell growth by targeting specific factors or cell pathways associated with cell proliferation. However, the research on exosomal lncRNAs and tumor growth is far more abundant109,110,111.
Exosomal lncRNAs in tumor metastasis
Recently, research on exosomal lncRNAs has made rapid progress. For example, Jin et al. suggested that the lncRNA TIRY was able to promote tumor metastasis by enhancing the EMT process in oral cancer112. Similarly, Q Zhang et al. demonstrated that SKOV3-secreted exosomes suppressed the phosphatase and tensin homolog (PTEN)/AKT signaling pathway through the transfer of the lncRNA FAL1, thereby inhibiting OC cell metastasis in vitro and in vivo113. In addition, in a study of ESCC progression, Li et al. found that ESCC tissues showed high expression of lncRNA-ZFAS1 and low expression of miR-124. Gene silencing experiments confirmed that the silencing of ZNFX1 antisense RNA 1 (ZFAS1) inhibited the development of ESCC79. Additionally, exosomal lncRNA-MALAT1 was found to be upregulated in NSCLC114. In vitro studies have shown that exosomal lncRNA-MALAT1 facilitates tumor proliferation and metastasis and inhibits tumor apoptosis. Mechanistically, it mainly exerts its function by modulating the levels of cyclin D1 and cyclin D2 in NSCLC. In contrast, exosomal lncRNA-PTENP1, an exosome with reduced expression in bladder cancer patients, was shown to inhibit the biological malignancy of bladder cancer cells by increasing apoptosis rates and reducing cell invasion and migration capacity115. Not surprisingly, other similar studies have revealed the role of exosomal lncRNAs in tumor metastasis116,117,118,119.
Exosomal lncRNAs in tumor angiogenesis
An increasing number of studies have shown that unusual lincRNA expression has an important effect on tumor progression, including angiogenesis and tumor development120. Conigliaro et al. showed that exosomes that are high in lincRNA H19 secreted by CD90+ LCCs could be absorbed by endothelial cells and promote angiogenic phenotypes and intercellular adhesion121. In terms of the mechanism, functional studies have demonstrated that exosomal lincRNA H19 functions by managing the levels of VEGF and VEGF receptor 1 (VEGFR1). Dynamic changes in endothelial cells are an important feature of angiogenesis122. However, many genes are involved in endothelial cell function123. The study of Lang et al. suggested that exosomal lincRNA-POU3F3 was abnormally elevated in glioma and was associated with advanced tumors. Further mechanistic studies revealed that glioma cells were able to transport lincRNA-POU3F3 to endothelial cells in the form of exosomes and induce angiogenesis by modulating angiogenic factor levels124. Additionally, in another study of glioma, exosomes from U87-MG cells that were enriched in lincRNA-CCAT2 were also shown to be internalized by HUVECs, thereby promoting the angiogenesis of HUVECs125. Furthermore, in lung cancer cells, the exosomal lncRNA GAS5 could bind to miRNA-29-3P in competition with PTEN, thereby inhibiting phosphatidylinositol 3-kinase (PI3K) levels and promoting HUVEC proliferation and tube formation126. In general, tumor cells are able to transfer lncRNAs to endothelial cells through exosomes, and these lncRNAs can promote the production of angiogenesis-related factors and molecules, thus promoting tumor angiogenesis.
Exosomal lncRNAs in tumor chemoresistance
Recently, the relationship between exosomal lncRNAs and tumor chemoresistance has received increasing attention. A study by Wang et al. showed that exosomal lncRNA H19 can be delivered to doxorubicin-sensitive BC cells, leading to the dissemination of doxorubicin resistance127. In another study on BC, a significant increase in exosomal lncRNA-SNHG1 was found in trastuzumab-resistant cells128. Functional experimentation showed that lncRNA-SNHG14 knockdown could effectively promote the efficacy of trastuzumab. According to the Signal Transduction Reporter Array, the lncRNA SNHG14 was likely to generate resistance by targeting the apoptosis regulator Bcl-2/Bax signaling pathways. Likewise, the expression levels of lncRNA-UCA1 were notably upregulated in BCC patients with tamoxifen resistance129.
In addition to BCCs, exosomal lncRNAs have also been extensively studied in other tumors. For example, the exosomal lncRNA HOTTIP was shown to contribute to the cisplatin resistance of GC cells by regulating the miR-218/high mobility group AT-hook 1 (HMGA1) axis130. Additionally, in a study of erlotinib resistance in NSCLC, Wei Zhang et al. revealed that in erlotinib-resistant NSCLC cells, the expression of the exosomal lncRNA RP11-838N2.4 was elevated131. Likewise, in a study on ESCC resistance, Min Kang et al. demonstrated that the levels of the exosomal lncRNA PART1 were upregulated in ESCC cells, which generated gefitinib resistance. Experimental results have shown that the lncRNA PART1 promotes the generation of resistance by regulating the miR-129/Bcl-2 pathway132. For renal cell carcinoma (RCC), lncRNA profile analysis and sequence screening indicated that lncRNA-ARSR was highly enriched in sunitinib-resistant RCC cells. Further experiments showed that lncRNA-ARSR promoted RCC cell resistance to sunitinib by competitively binding miR-34/miR-449 to promote AXL receptor tyrosine kinase (AXL) and c-MET expression133. However, more studies on the relationship between exosomal lncRNAs and chemoresistance are ongoing, which will help us better understand the mechanism of tumor resistance134,135,136.
Other exosomal ncRNAs in cancer
snRNAs
snRNAs are the major component of RNA spliceosomes in eukaryotic posttranscriptional processing and participate in the processing of mRNA precursors. Endogenous variation in snRNAs could potentially enable these RNAs to regulate alternative splicing to drive genetic, dysplastic, and neoplastic disease137,138 However, there are a very limited number of studies exploring the roles of exosomal snRNAs in cancer progression. One study revealed that exosomal snRNAs in cancer cells mediate the activation of Toll-like receptor 3, therefore inducing chemokine secretion, which contributes to neutrophil mobilization and recruitment, leading to the generation of the lung premetastatic niche and further metastasis139. In addition, another recent study suggested that exosomal snRNA U1, U2, and U5 levels were significantly decreased in patients with lung cancer compared with early-stage patients, indicating their potential as biomarkers in lung cancer140. Nevertheless, the function of secreted snRNAs in cancer has largely not been explored, and more attention should be given to this topic.
snoRNAs
snoRNAs are a class of small molecular ncRNAs widely distributed in the nucleolus of eukaryotic cells. They have conserved structural elements and can be divided into three categories: box C/D snoRNA, box H/ACA snoRNA, and MRPRNA. SnoRNAs have a significant influence on the posttranscriptional modification of rRNA. However, few studies on exosomal snoRNAs in cancer have been reported to date141. Only one study revealed that the snoRNAs SNORD33, SNORD66 and SNORD76 showed higher expression levels in NSCLC patients than in healthy individuals142. Such a situation raises many questions, such as what roles do snoRNAs play in cancer and how do they function? We believe that an in-depth understanding of exosomal snoRNA in tumors may contribute to the design of cancer-diagnostic and cancer-prognostic tools. Obviously, more attention should be given to this subject.
Clinical implications of exosomal ncRNAs in cancer treatment
The poor prognosis of some cancer treatments is largely due to the detection of late-stage disease, partly caused by the deficiency of suitable biomarkers. As discussed above, exosomal ncRNAs have a key effect on the pathological process of various cancers, thus exosomal ncRNAs associated with the generation and progression of tumors can potentially be used as diagnostic, prognostic, and predictive biomarkers for cancer. In addition, exosomes are present in a variety of body fluids, such as blood, urine, saliva, breast milk, and ascites143,144,145,146,147, and this makes it easy to collect them and detect their levels. Therefore, they can be used as a liquid tool for noninvasive clinical testing.
To date, the most widely studied exosomal ncRNAs are exosomal miRNAs. Multiple studies have shown that exosomal miRNAs have the potential to be used as tools for the diagnosis of various cancers. For example, a study revealed the underlying role of exosomal miR-373 as a biomarker for aggressive tumors. Its identification can be used for the diagnosis of BC53. In another study, miRNA expression profiles were conducted to identify exosomal miRNAs that could distinguish malignant cancer from benign disease. The results showed that eight tumor-specific exosomal miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205, and miR-214) were significantly distinct in profiles between benign disease and OC, and they could be used as surrogate diagnostic markers for biopsy profiling148. Moreover, Cazzoli et al. suggested that several exosomal miRNAs (miR‐200b-5p, miR-378a, miR-139-5p, and miR-379) could distinguish nodules and nonnodules and that other exosomal miRNAs (miR-629, miR-30a-3p, miR-100, miR-200b-5p, miR-154-3p, and miR-151a-5p) could discriminate granulomas from lung adenocarcinoma, indicating their diagnostic value in lung cancer149. Of course, in addition to being diagnostic biomarkers, exosomal miRNAs have also been shown to be used as prognostic biomarkers for cancer. In a study by Xiaoyi Huang et al., it was shown that high levels of two plasma exosomal miRNAs (miR-1290 and miR-375) were significantly associated with poor overall survival in castration-resistant PCa, indicating their potential as prognostic biomarkers150. Similarly, compared to primary tumors, exosomal miR-21 and miR-155 were significantly increased in recurrent tumors, which suggested their potential as prognostic biomarkers151. Interestingly, exosomal miRNA-21 has also been shown to be related to the TNM stage of HCC, and its high level was associated with poor prognosis in CRC152,153. These distinct exosomal miRNAs function by regulating differential signaling pathways and may serve as promising targets for cancer therapy.
Apart from exosomal miRNAs, some researchers have suggested that exosomal lncRNAs could act as a diagnostic tool in cancer. A study showed that the levels of the exosomal lncRNAs ENSG00000258332.1 and LINC00635 were significantly increased in HCC patients compared with chronic hepatitis B patients154. Studies have shed light on lncRNAs from exosomes as diagnostic biomarkers for HCC cells. Similarly, an analysis demonstrated that higher expression levels of exosomal lncRNA ATB can serve as a prediction tool in cancer prognosis, since the overall survival of patients in the exosomal lncRNA ATB high expression groups was shorter152. Another study indicated that elevated levels of the lncRNA MIAT were related to shorter survival in GC patients155. Recently, based on public databases and the integration of bioinformatics analyses, exosomal lncRNA BCRT1 was demonstrated to be a potential biomarker and therapeutic target for BC108.
Compared with other ncRNAs that exhibit the potential to function as cancer biomarkers, circRNAs with a more stable circular structure seem to have more potential as tumor biomarkers. A study showed that the increased expression of exosomal circPRMT5 was associated with advanced clinical stage and poor survival in patients with urothelial carcinoma of bladder cancer patients, and data demonstrated that the exosomal circRNA PRMT5 exerts its protumor effect by sponging miR-30c. An analysis suggested that the exosomal circRNA PRMT5 may act as an important therapeutic target for patients156. In another study, exosomal circRNA FNDC3B levels were elevated in papillary thyroid carcinoma (PTC) tissues and cell lines. Guojun Wu et al. showed that the exosomal circRNA FNDC3B regulated PTC progression via the miR-1178/TLR4 axis and suggested that it was likely to be a potential target for the treatment of PTC157. Furthermore, hsa_circ_0000419, hsa_circ_0065149, and circ-KIAA1244 were suggested to serve as potential diagnostic and/or prognostic biomarkers for GC158,159,160. Exosomal circRNAs represent promising biomarkers and therapeutic targets in cancer. Nonetheless, the study of the role of exosomal circRNAs in cancer is still ongoing.
However, for exosomal snRNAs, only one study revealed that the decreased levels of exosomal snRNA U1, U2 and U5 were correlated with lung cancer progression, possessing favorable diagnostic efficiency, suggesting that the aberrant expression of snRNA U1, U2, and U5 could act as novel biomarkers for lung cancer140.
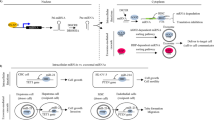
With developments in biotechnology, exosomes can replace synthetic nanoparticles. As natural nanoparticles, exosomes have better biocompatibility and lower biological toxicity and immunogenicity; therefore, exosomal ncRNAs are a potential choice for novel cancer treatments. A study has shown that synthesized sphingosine kinase 2 (Sphk2) siRNA-loaded nanoparticles could reduce exosomal miR-21 levels and inhibit tumor migration in HCC cells161. In addition, Li et al. used BMSCs to obtain modified exosomes to deliver GRP78 siRNA to HCC. The results showed that GRP78 siRNA combined with sorafenib can target glucose-regulated protein 78 (GRP78), which is overexpressed in sorafenib-resistant HCC and sensitizes sorafenib-resistant HCC to sorafenib, thus reversing drug resistance162. In addition, Chunhui Wang et al. found that transient receptor potential polycystic 2 (TRPP2) expression was increased in laryngeal squamous cell carcinoma (LSCC) and promoted the metastatic ability of cancer cells by regulating the EMT process. By targeting TRPP2 with the exosome/TRPP2 siRNA complex, the expression of N-cadherin and Vimentin in LSCC cells was notably reduced, while the expression of E-cadherin was elevated. This revealed that the exosome/TRPP2 siRNA complex could effectively inhibit the metastasis of LSCC163. These studies reveal a novel cancer therapeutic method involving the ablation of oncogenic miRNAs from exosomes.
Generally, exosomal ncRNAs have great prospects in the applications of cancer therapy due to their structural stability, which effectively prevents the digestion and degradation of RNA by related enzymes. Given the biocompatibility of exosomes, they can be engineered to deliver therapeutic factors (such as RNA, proteins and drugs) to target cells for therapeutic applications. In addition, based on the targeting of exosome transport, they can be used as carriers of drugs in vivo, which has great potential in clinical applications such as targeted therapy for tumors and disease course monitoring (Fig. 3).
Discussion
Exosomes exert their vital functions in many pathophysiological processes, such as inflammatory reactions, immunoregulation, tumor progression, tumor invasion, and metastasis. NcRNAs exist in different intercellular compartments and participate in different cellular reactions. They can also be identified in biological fluids and are called circulating ncRNAs, and these ncRNAs can be detected in exosomes.
Specifically, in this review, we highlighted the biological characteristics of exosomes and presented a comprehensive update on the functions of exosomal miRNAs, circRNAs, and lncRNAs in tumor growth, metastasis, angiogenesis and tumor chemoresistance. The role of these ncRNAs may be tumor supportive or tumor suppressive in different cancers. Therefore, considerable focus has shifted to the cancer-related signatures of these ncRNAs.
We also described the therapeutic implications of exosomal ncRNAs in cancer treatment, suggesting that many ncRNAs may be regarded as predictive markers. Due to abnormal chromatin remodeling and alternative splicing programs in cancer, lncRNAs and circRNAs can be extremely cancer type-specific. Thus, exosomal lncRNAs and circRNAs can be useful cancer-specific biomarkers. Because exosomal ncRNAs have great diagnostic and therapeutic potential, a large number of studies indicate that further exploration is needed, but there are still many challenges. More research on the roles and regulation of the identified ncRNAs is needed to understand their molecular mechanism.
References
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Cocozza, F., Grisard, E., Martin-Jaular, L., Mathieu, M. & Théry, C. SnapShot: extracellular vesicles. Cell 182, 262–262.e261 (2020).
Greening, D. W. et al. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin. Cell Dev. Biol. 40, 60–71 (2015).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Théry, C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15 (2011).
Iero, M. et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 15, 80–88 (2008).
Lee, Y. J. et al. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials 101, 143–155 (2016).
Zhao, Y. M. et al. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer 118, 2708–2717 (2012).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750–1535750 (2018).
Théry, C., Zitvogel, L. & Amigorena, S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 (2002).
Trams, E. G., Lauter, C. J., Salem, N. Jr. & Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 645, 63–70 (1981).
Harding, C., Heuser, J. & Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 97, 329–339 (1983).
Johnstone, R. M., Adam, M., Hammond, J. R., Orr, L. & Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420 (1987).
Cabral, J., Ryan, A. E., Griffin, M. D. & Ritter, T. Extracellular vesicles as modulators of wound healing. Adv. Drug Deliv. Rev. 129, 394–406 (2018).
Lindenbergh, M. F. S. & Stoorvogel, W. Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu. Rev. Immunol. 36, 435–459 (2018).
Becker, A. et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 30, 836–848 (2016).
György, B. et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 68, 2667–2688 (2011).
de Gassart, A., Géminard, C., Hoekstra, D. & Vidal, M. Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic 5, 896–903 (2004).
Kharaziha, P., Ceder, S., Li, Q. & Panaretakis, T. Tumor cell-derived exosomes: a message in a bottle. Biochim. Biophys. Acta 1826, 103–111 (2012).
Cai, Z. et al. Activated T cell exosomes promote tumor invasion via Fas signaling pathway. J. Immunol. 188, 5954–5961 (2012).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 32, 623–642 (2013).
Huang, X. et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14, 319 (2013).
Cheng, L., Sun, X., Scicluna, B. J., Coleman, B. M. & Hill, A. F. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 86, 433–444 (2014).
Liu, T. et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 7, 85551–85563 (2016).
Lim, L. P. et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 (2005).
Calin, G. A. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353, 1793–1801 (2005).
Umezu, T., Ohyashiki, K., Kuroda, M. & Ohyashiki, J. H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 32, 2747–2755 (2013).
Zhang, Y. et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 39, 133–144 (2010).
Tkach, M. & Théry, C. Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232 (2016).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017).
Xu, W. W. et al. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat. Commun. 8, 14399 (2017).
Li, B. et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene 36, 3986–4000 (2017).
Chiam, K. et al. Circulating serum exosomal miRNAs As potential biomarkers for esophageal adenocarcinoma. J. Gastrointest. Surg. 19, 1208–1215 (2015).
Kogure, T., Lin, W. L., Yan, I. K., Braconi, C. & Patel, T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 54, 1237–1248 (2011).
Grimolizzi, F. et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci. Rep. 7, 15277 (2017).
Monaco, F. et al. Exosomal transfer of miR-126 promotes the anti-tumour response in malignant mesothelioma: Role of miR-126 in cancer-stroma communication. Cancer Lett. 463, 27–36 (2019).
Wang, L., He, M., Fu, L. & Jin, Y. Exosomal release of microRNA-454 by breast cancer cells sustains biological properties of cancer stem cells via the PRRT2/Wnt axis in ovarian cancer. Life Sci. 257, 118024 (2020).
Melo, S. A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014).
Fang, T. et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 9, 191 (2018).
Zhou, W. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515 (2014).
Fong, M. Y. et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194 (2015).
Matsumura, T. et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 113, 275–281 (2015).
Bryant, R. J. et al. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 106, 768–774 (2012).
Li, Z. et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 9, 139–148 (2016).
Nishida, N., Yano, H., Nishida, T., Kamura, T. & Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag 2, 213–219 (2006).
Zhuang, G. et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 31, 3513–3523 (2012).
Cui, H. et al. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene 34, 3640–3650 (2015).
Kosaka, N. et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 288, 10849–10859 (2013).
Liu, Y. et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 370, 125–135 (2016).
Wu, X. G. et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis 22, 397–410 (2019).
Zheng, H. C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 8, 59950–59964 (2017).
Eichelser, C. et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 5, 9650–9663 (2014).
Challagundla, K. B. et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 107, 7 (2015).
Sasako, M. et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J. Clin. Oncol. 29, 4387–4393 (2011).
Sakuramoto, S. et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N. Engl. J. Med. 357, 1810–1820 (2007).
Tsuburaya, A. et al. Sequential paclitaxel followed by tegafur and uracil (UFT) or S-1 versus UFT or S-1 monotherapy as adjuvant chemotherapy for T4a/b gastric cancer (SAMIT): a phase 3 factorial randomised controlled trial. Lancet Oncol. 15, 886–893 (2014).
Jin, G. et al. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 84, 315–325 (2019).
Li, B. et al. Identification of miR-29c and its Target FBXO31 as a key regulatory mechanism in esophageal cancer chemoresistance: functional validation and clinical significance. Theranostics 9, 1599–1613 (2019).
Zhou, Y. et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 37, 324 (2018).
Olejarz, W., Kubiak-Tomaszewska, G., Chrzanowska, A. & Lorenc, T. Exosomes in Angiogenesis and Anti-angiogenic Therapy in Cancers. Int. J. Mol. Sci. 21, 16 (2020).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Ørom, U. A., Nielsen, F. C. & Lund, A. H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471 (2008).
Kulkarni, B. et al. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discov. Today 24, 2058–2067 (2019).
Zhao, S. et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 13, 156 (2020).
Zhang, X. et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 18, 40 (2019).
Lin, Q. et al. Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am. J. Transl. Res. 12, 1080–1095 (2020).
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B. & Kahn, C. R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673 (2019).
Yan, W. et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 20, 597–609 (2018).
Zheng, P. et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 36, 53 (2017).
Lu, J. et al. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 37, 147 (2018).
Huang, S. et al. The emerging role of circular RNAs in transcriptome regulation. Genomics 109, 401–407 (2017).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Fanale, D., Taverna, S., Russo, A. & Bazan, V. Circular RNA in exosomes. Adv. Exp. Med. Biol. 1087, 109–117 (2018).
Li, Y. et al. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin. Sci. 133, 2463–2479 (2019).
Feng, W. et al. circIFT80 Functions as a ceRNA of miR-1236-3p to Promote Colorectal Cancer Progression. Mol. Ther. Nucleic Acids 18, 375–387 (2019).
Zhang, H. et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 38, 2844–2859 (2019).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Zhang, X. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 18, 20 (2019).
Xie, M. et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 19, 112 (2020).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Wang, J. et al. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging 12, 11550–11567 (2020).
Li, L. et al. FLI1 exonic circular rnas as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 25, 1302–1317 (2019).
Xu, L. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 143, 17–27 (2017).
Yang, H. et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics 10, 8211–8226 (2020).
Huang, X. Y. et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 39, 20 (2020).
Geng, X. et al. Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 12, 267–283 (2020).
Ding, C. et al. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 479, 1–12 (2020).
Wang, X. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 14, 539–555 (2020).
Hon, K. W., Ab-Mutalib, N. S., Abdullah, N. M. A., Jamal, R. & Abu, N. Extracellular Vesicle-derived circular RNAs confers chemoresistance in colorectal cancer. Sci. Rep. 9, 16497 (2019).
Zhang, P. F. et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 19, 110 (2020).
Li, Z. et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 432, 237–250 (2018).
Li, J. et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 37, 177 (2018).
Dai, X. et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 9, 454 (2018).
Chen, W. et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 475, 119–128 (2020).
Wang, G. et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 40, 432–445 (2019).
Wang, S. et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin. Sci. 133, 1935–1953 (2019).
Zong, Z. H., Du, Y. P., Guan, X., Chen, S. & Zhao, Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 38, 437 (2019).
Wang, R. et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res 78, 4812–4825 (2018).
Tian, L. et al. CircRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin. Sci. 133, 1053–1066 (2019).
Dou, Y. et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 6, 37982 (2016).
Moran, V. A., Perera, R. J. & Khalil, A. M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40, 6391–6400 (2012).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Zhang, Y. et al. Exosome-Transmitted lncRNA H19 Inhibits the Growth of Pituitary Adenoma. J. Clin. Endocrinol. Metab. 104, 6345–6356 (2019).
Yang, X. et al. The long non-coding RNA PCSEAT exhibits an oncogenic property in prostate cancer and functions as a competing endogenous RNA that associates with EZH2. Biochem. Biophys. Res. Commun. 502, 262–268 (2018).
Liang, Y. et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 19, 85 (2020).
Zang, X. et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 11, 215 (2020).
Li, B. et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene 37, 5508–5519 (2018).
Xue, M. et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer 16, 143 (2017).
Jin, N. et al. Long non-coding RNA TIRY promotes tumor metastasis by enhancing epithelial-to-mesenchymal transition in oral cancer. Exp. Biol. Med. 245, 585–596 (2020).
Zhang, Q., Len, T. Y., Zhang, S. X., Zhao, Q. H. & Yang, L. H. Exosomes transferring long non-coding RNA FAL1 to regulate ovarian cancer metastasis through the PTEN/AKT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 43–54 (2020).
Zhang, R. et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 490, 406–414 (2017).
Zheng, R. et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol. Cancer 17, 143 (2018).
Hardin, H. et al. Thyroid cancer stem-like cell exosomes: regulation of EMT via transfer of lncRNAs. Lab. Investig. 98, 1133–1142 (2018).
Wu, D. M. et al. TGF-β-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 7, 5118–5129 (2018).
Gao, T. et al. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 18, 11 (2018).
Liang, Z. X. et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 10, 829 (2019).
Meseure, D., Drak Alsibai, K., Nicolas, A., Bieche, I. & Morillon, A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed. Res. Int. 2015, 320214 (2015).
Conigliaro, A. et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 14, 155 (2015).
Herbert, S. P. & Stainier, D. Y. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 551–564 (2011).
Zhao, Y. & Adjei, A. A. Targeting angiogenesis in cancer therapy: moving beyond vascular endothelial growth factor. oncologist 20, 660–673 (2015).
Lang, H. L. et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 21, 959–972 (2017).
Lang, H. L. et al. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 38, 785–798 (2017).
Cheng, Y. et al. Low long noncoding RNA growth arrest-specific transcript 5 expression in the exosomes of lung cancer cells promotes tumor angiogenesis. J. Oncol. 2019, 2476175 (2019).
Wang, X. et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 235, 6896–6904 (2020).
Dong, H. et al. Exosome-mediated transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 53, 1013–1026 (2018).
Xu, C. G., Yang, M. F., Ren, Y. Q., Wu, C. H. & Wang, L. Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 20, 4362–4368 (2016).
Wang, J. et al. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther. 12, 11325–11338 (2019).
Zhang, W. et al. Exosome-mediated transfer of lncRNA RP11‑838N2.4 promotes erlotinib resistance in non-small cell lung cancer. Int. J. Oncol. 53, 527–538 (2018).
Kang, M. et al. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J. Exp. Clin. Cancer Res. 37, 171 (2018).
Qu, L. et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668 (2016).
Takahashi, K., Yan, I. K., Wood, J., Haga, H. & Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 12, 1377–1387 (2014).
Takahashi, K., Yan, I. K., Kogure, T., Haga, H. & Patel, T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 4, 458–467 (2014).
Luo, X. et al. Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell Int. 19, 323 (2019).
Dvinge, H., Guenthoer, J., Porter, P. L. & Bradley, R. K. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 29, 1591–1604 (2019).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Liu, Y. et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016).
Dong, X. et al. Small nuclear RNAs (U1, U2, U5) in tumor-educated platelets are downregulated and act as promising biomarkers in lung cancer. Front. Oncol. 10, 1627–1627 (2020).
Scott, M. S. & Ono, M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie 93, 1987–1992 (2011).
Liao, J. et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 9, 198 (2010).
Morse, M. A. et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 3, 9 (2005).
Merchant, M. L., Rood, I. M., Deegens, J. K. J. & Klein, J. B. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat. Rev. Nephrol. 13, 731–749 (2017).
Zhou, Q. et al. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 8, 118–123 (2012).
Peng, P., Yan, Y. & Keng, S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol. Rep. 25, 749–762 (2011).
Li, M. et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 1652 (2014).
Taylor, D. D. & Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 (2008).
Cazzoli, R. et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 8, 1156–1162 (2013).
Huang, X. et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 67, 33–41 (2015).
Munagala, R., Aqil, F. & Gupta, R. C. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. 37, 10703–10714 (2016).
Lee, Y. R. et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 144, 1444–1452 (2019).
Tsukamoto, M., Iinuma, H., Yagi, T., Matsuda, K. & Hashiguchi, Y. Circulating exosomal MicroRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92, 360–370 (2017).
Xu, H., Chen, Y., Dong, X. & Wang, X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol. Biomark. Prev. 27, 710–716 (2018).
Xu, H. et al. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 34, e23323 (2020).
Chen, X. et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 24, 6319–6330 (2018).
Wu, G. et al. Circular RNA profiling reveals exosomal circ_0006156 as a novel biomarker in papillary thyroid cancer. Mol. Ther. Nucleic Acids 19, 1134–1144 (2020).
Tao, X. et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol. Res. Pract. 216, 152763 (2020).
Shao, Y. et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol. Oncol. Res. 26, 1475–1482 (2020).
Tang, W. et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 17, 137 (2018).
Liang, J. et al. Sphk2 RNAi nanoparticles suppress tumor growth via downregulating cancer cell derived exosomal microRNA. J. Control. Release 286, 348–357 (2018).
Li, H., Yang, C., Shi, Y. & Zhao, L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J. Nanobiotechnology 16, 103 (2018).
Wang, C. et al. Exosome-delivered TRPP2 siRNA inhibits the epithelial-mesenchymal transition of FaDu cells. Oncol. Lett. 17, 1953–1961 (2019).
Funding
This work was supported by the National Natural Sciences Foundation of China (82073196, 81803551), Guangdong Natural Science Research Grant International joint project (2021A0505030035), Guangdong Natural Science Research Grant (2021A1515011158), and the Fundamental Research Funds for the Central Universities (21620429).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All the authors agree to publish this paper.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, QW., He, Y. & Xu, W.W. Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer. Exp Mol Med 54, 216–225 (2022). https://doi.org/10.1038/s12276-022-00744-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-022-00744-w
This article is cited by
-
Circular RNAs in tumor immunity and immunotherapy
Molecular Cancer (2024)
-
LncRNA LUESCC promotes esophageal squamous cell carcinoma by targeting the miR-6785-5p/NRSN2 axis
Cellular and Molecular Life Sciences (2024)
-
Identification of exosomal microRNA panel as diagnostic and prognostic biomarker for small cell lung cancer
Biomarker Research (2023)
-
CircUSP48 promotes malignant behavior by regulating CYR61 via miR-365 in osteosarcoma
Functional & Integrative Genomics (2023)
-
Exosomal microRNAs and long noncoding RNAs: as novel biomarkers for endometriosis
Cell and Tissue Research (2023)