Abstract
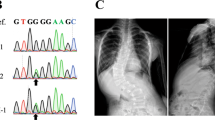
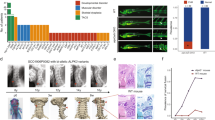
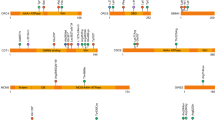
Spondylocostal dysostosis (SCDO) encompasses a group of skeletal disorders characterized by multiple segmentation defects in the vertebrae and ribs. SCDO has a complex genetic etiology. This study aimed to analyze and identify pathogenic variants in a fetus with SCDO. Copy number variant sequencing and whole exome sequencing were performed on a Chinese fetus with SCDO, followed by bioinformatics analyses, in vitro functional assays and a systematic review on the reported SCDO cases with LFNG pathogenic variants. Ultrasound examinations in utero exhibited that the fetus had vertebral malformation, scoliosis and tethered cord, but rib malformation was not evident. We found a novel homozygous variant (c.1078 C > T, p.R360C) within the last exon of LFNG. The variant was predicted to cause loss of function of LFNG by in silico prediction tools, which was confirmed by an in vitro assay of LFNG enzyme activity. The systematic review listed a total of 20 variants of LFNG in SCDO. The mutational spectrum spans across all exons of LFNG except the last one. This study reported the first Chinese case of LFNG-related SCDO, revealing the prenatal phenotypes and expanding the mutational spectrum of the disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Solomon L, Jimenez RB, Reiner L. Spondylothoracic dysostosis: report of two cases and review of the literature. Arch Pathol Lab Med. 1978;102:201–5.
Gucev ZS, Tasic V, Pop-Jordanova N, Sparrow DB, Dunwoodie SL, Ellard S, et al. Autosomal dominant spondylocostal dysostosis in three generations of a Macedonian family: negative mutation analysis of DLL3, MESP2, HES7, and LFNG. Am J Med Genet A. 2010;152A:1378–82. https://doi.org/10.1002/ajmg.a.33471
Whittock NV, Sparrow DB, Wouters MA, Sillence D, Ellard S, Dunwoodie SL, et al. Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet. 2004. https://doi.org/10.1086/421053
Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, et al. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet. 2006;78:28–37. https://doi.org/10.1086/498879
Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL. Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet. 2008;17:3761–6. https://doi.org/10.1093/hmg/ddn272
Umair M, Younus M, Shafiq S, Nayab A, Alfadhel M. Clinical genetics of spondylocostal dysostosis: a mini review. Front Genet. 2022;13:996364. https://doi.org/10.3389/fgene.2022.996364
Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–50. https://doi.org/10.1056/NEJMoa1406829
McInerney-Leo AM, Sparrow DB, Harris JE, Gardiner BB, Marshall MS, O’Reilly VC, et al. Compound heterozygous mutations in RIPPLY2 associated with vertebral segmentation defects. Hum Mol Genet. 2015;24:1234–42. https://doi.org/10.1093/hmg/ddu534
Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, Lander ES, et al. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet. 2000;24:438–41. https://doi.org/10.1038/74307
Turnpenny PD, Whittock N, Duncan J, Dunwoodie S, Kusumi K, Ellard S. Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J Med Genet. 2003;40:333–9. https://doi.org/10.1136/jmg.40.5.333
Zhang S, Qiao Y, Wang Z, Zhuang J, Sun Y, Shang X, et al. Identification of novel compound heterozygous variants in SLC19A2 and the genotype-phenotype associations in thiamine-responsive megaloblastic anemia. Clin Chim Acta. 2021;516:157–68. https://doi.org/10.1016/j.cca.2021.01.025
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. https://doi.org/10.1038/nmeth0410-248
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. https://doi.org/10.1038/nmeth0810-575
Shangguan H, Su C, Ouyang Q, Cao B, Wang J, Gong C, et al. Kabuki syndrome: novel pathogenic variants, new phenotypes and review of literature. Orphanet J Rare Dis. 2019;14:255. https://doi.org/10.1186/s13023-019-1219-x
Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–5. https://doi.org/10.1093/nar/gky300
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–82. https://doi.org/10.1038/s41592-022-01488-1
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med Off J Am Coll Med Genet. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30
Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, et al. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem. 2005;280:42454–63. https://doi.org/10.1074/jbc.M509552200
Otomo N, Mizumoto S, Lu HF, Takeda K, Campos-Xavier B, Mittaz-Crettol L, et al. Identification of novel LFNG mutations in spondylocostal dysostosis. J Hum Genet. 2019;64:261–4. https://doi.org/10.1038/s10038-018-0548-2
Okubo Y, Sugawara T, Abe-Koduka N, Kanno J, Kimura A, Saga Y. Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of Notch signalling. Nat Commun. 2012;3:1141. https://doi.org/10.1038/ncomms2133
Bochter MS, Servello D, Kakuda S, D’Amico R, Ebetino MF, Haltiwanger RS, et al. Lfng and Dll3 cooperate to modulate protein interactions in cis and coordinate oscillatory Notch pathway activation in the segmentation clock. Dev Biol. 2022;487:42–56. https://doi.org/10.1016/j.ydbio.2022.04.004
Matsumoto K, Kumar V, Varshney S, Nairn AV, Ito A, Pennarubia F, et al. Fringe GlcNAc-transferases differentially extend O-fucose on endogenous NOTCH1 in mouse activated T cells. J Biol Chem. 2022;298:102064. https://doi.org/10.1016/j.jbc.2022.102064
Xu K, Nieuwenhuis E, Cohen BL, Wang W, Egan SE. Lunatic fringe-mediated Notch signaling is required for lung alveogenesis. Am J Physiol Lung Cell Mol Physiol. 2010;298:L45–56. https://doi.org/10.1152/ajplung.90550.2008
Acknowledgements
We thank the patients and their families for their help to the study.
Funding
This study was supported by Key Research & Develop program of Shaanxi Province (Grant nos. 2023-YBSF-305), Interior Program in Northwest Women’s and Children’s Hospital (2022YN24), Supporting Program of Innovation Capability Shaanxi Province (2024CX-GXPT-40, L.G.), Young Talent Support Plan from Xi’an Jiaotong University (YX6J033, L.G.), Provincial Talent Research Support Fund (71240000000131, L.G.), a Grant-in-Aid for Scientific Research (C) 23K06142 (to S.M.) from the Japan Society for the Promotion of Science, Pathogenesis of Intractable Diseases from the Research Institute of Meijo University (S.M. and S.Y.), Health Research Foundation of ShaanXi Province (Grant No. 2022D058), National Natural Science Foundation of China (Grant No. 82201865), and Basic scientific research funds of Xi’an Jiaotong University (Grant No. xzy012021074).
Author information
Authors and Affiliations
Contributions
Long Guo and Rong Qiang designed the study. Lin Wang, Shuji Mizumoto, Shuhei Yamada, Ruixue Zhang, Xin Li, Wenjing Cheng, Jiaqi Han, Lianying Jiao, Yating Wang, Qiujie Jin, Lihui Yang carried out the research. Long Guo, Lin wang, Shuji Mizumoto, Shuhei Yamada, Ruixue Zhang, Min Dan, Chunyan Zhang, Xinru Gao, Juan Wang, Chenxing Li, Jinhui Zhu, Shuxian Li, Hai Jiang, Gen Nishimura, Takahiro Yamada, Na Cai analyzed and interpreted data. Lin Wang drafted the manuscript. Long Guo, Rong Qiang revised it. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The research project was approved by Ethics Committee of Northwest Women’s and Children’s Hospital (No. 21-036).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Mizumoto, S., Zhang, R. et al. Identification of a novel LFNG variant in a Chinese fetus with spondylocostal dysostosis and a systematic review. J Hum Genet (2024). https://doi.org/10.1038/s10038-024-01248-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s10038-024-01248-3