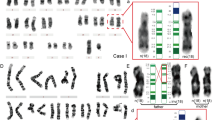
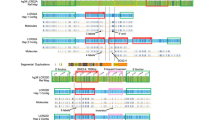
Abstract
Complex chromosomal rearrangements (CCRs) can result in spontaneous abortions, infertility, and malformations in newborns. In this study, we explored a familial CCR involving chromosome 6 by combining optical genomic mapping (OGM) and molecular cytogenetic methodologies. Within this family, the father and the paternal grandfather were both asymptomatic carriers of an identical balanced CCR, while the two offspring with an unbalanced paternal-origin CCR and two microdeletions presented with clinical manifestation. The first affected child, a 5-year-old boy, exhibited neurodevelopmental delay, while the second, a fetus, presented with hydrops fetalis. SNP-genotype analysis revealed a recombination event during gamete formation in the father that may have contributed to the deletion in his offspring. Meanwhile, the couple’s haplotypes will facilitate the selection of normal gametes in the setting of assisted reproduction. Our study demonstrated the potential of OGM in identifying CCRs and its ability to work with current methodologies to refine precise breakpoints and construct accurate haplotypes for couples with a CCR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data presented in this study are available on request from the corresponding author.
References
Patsalis PC. Complex chromosomal rearrangements. Genet Couns. 2007;18:57–69.
Madan K. What is a complex chromosome rearrangement? Am J Med Genet A. 2013;161a:1181–4.
Madan K, Nieuwint AW, van Bever Y. Recombination in a balanced complex translocation of a mother leading to a balanced reciprocal translocation in the child. Review of 60 cases of balanced complex translocations. Hum Genet. 1997;99:806–15.
Pai GS, Thomas GH, Mahoney W, Migeon BR. Complex chromosome rearrangements. Report of a new case and literature review. Clin Genet. 1980;18:436–44.
Sinkar P, Devi SR. Complex Chromosomal Rearrangement: A Case Report to Emphasize the Need for Parental Karyotyping and Genetic Counseling. J Hum Reprod Sci. 2020;13:68–70.
Ou J, Yang C, Cui X, Chen C, Ye S, Zhang C, et al. Successful pregnancy after prenatal diagnosis by NGS for a carrier of complex chromosome rearrangements. Reprod Biol Endocrinol. 2020;18:15.
Michaelson-Cohen R, Murik O, Zeligson S, Lobel O, Weiss O, Picard E, et al. Combining cytogenetic and genomic technologies for deciphering challenging complex chromosomal rearrangements. Mol Genet Genomics. 2022;297:925–33.
Giardino D, Corti C, Ballarati L, Colombo D, Sala E, Villa N, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn. 2009;29:257–65.
Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32.
Madan K. Balanced complex chromosome rearrangements: reproductive aspects. A review. Am J Med Genet A. 2012;158a:947–63.
Liao Y, Wang C, Liang M, Hu X, Wu Q. Analysis of genetic characteristics and reproductive risks of balanced complex chromosome rearrangement carriers in China. Hereditas (Beijing). 2017;39:396–412.
Pellestor F, Anahory T, Lefort G, Puechberty J, Liehr T, Hédon B, et al. Complex chromosomal rearrangements: origin and meiotic behavior. Hum Reprod Update. 2011;17:476–94.
Bayani J, Squire JA Fluorescence in situ Hybridization (FISH). Curr Protoc Cell Biol. 2004;Chapter 22:Unit 22.4.
Nowakowska BA, de Leeuw N, Ruivenkamp CA, Sikkema-Raddatz B, Crolla JA, Thoelen R, et al. Parental insertional balanced translocations are an important cause of apparently de novo CNVs in patients with developmental anomalies. Eur J Hum Genet. 2012;20:166–70.
Smeets DF. Historical prospective of human cytogenetics: from microscope to microarray. Clin Biochem. 2004;37:439–46.
Eisfeldt J, Pettersson M, Vezzi F, Wincent J, Käller M, Gruselius J, et al. Comprehensive structural variation genome map of individuals carrying complex chromosomal rearrangements. PLoS Genet. 2019;15:e1007858.
Sahajpal NS, Barseghyan H, Kolhe R, Hastie A, Chaubey A. Optical Genome Mapping as a Next-Generation Cytogenomic Tool for Detection of Structural and Copy Number Variations for Prenatal Genomic Analyses. Genes (Basel). 2021;12:398.
Zhang S, Pei Z, Lei C, Zhu S, Deng K, Zhou J, et al. Detection of cryptic balanced chromosomal rearrangements using high-resolution optical genome mapping. J Med Genet. 2023;60:274–84.
Yang Y, Hao W. Identification of a familial complex chromosomal rearrangement by optical genome mapping. Mol Cytogenet. 2022;15:41.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Burssed B, Zamariolli M, Bellucco FT, Melaragno MI. Mechanisms of structural chromosomal rearrangement formation. Mol Cytogenet. 2022;15:23.
Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49.
Salahshourifar I, Shahrokhshahi N, Tavakolzadeh T, Beheshti Z, Gourabi H. Complex chromosomal rearrangement involving chromosomes 1, 4 and 22 in an infertile male: case report and literature review. J Appl Genet. 2009;50:69–72.
Di Benedetto D, Di Vita G, Romano C, Giudice ML, Vitello GA, Zingale M, et al. 6p22.3 deletion: report of a patient with autism, severe intellectual disability and electroencephalographic anomalies. Mol Cytogenet. 2013;6:4.
Celestino-Soper PB, Skinner C, Schroer R, Eng P, Shenai J, Nowaczyk MM, et al. Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol Cytogenet. 2012;5:17.
Hamada N, Ogaya S, Nakashima M, Nishijo T, Sugawara Y, Iwamoto I, et al. De novo PHACTR1 mutations in West syndrome and their pathophysiological effects. Brain. 2018;141:3098–114.
Kiando SR, Tucker NR, Castro-Vega LJ, Katz A, D’Escamard V, Tréard C, et al. PHACTR1 is a genetic susceptibility locus for fibromuscular dysplasia supporting its complex genetic pattern of inheritance. PLoS Genet. 2016;12:e1006367.
Barøy T, Misceo D, Strømme P, Stray-Pedersen A, Holmgren A, Rødningen OK, et al. Haploinsufficiency of two histone modifier genes on 6p22.3, ATXN1 and JARID2, is associated with intellectual disability. Orphanet J Rare Dis. 2013;8:3.
Nazaryan-Petersen L, Eisfeldt J, Pettersson M, Lundin J, Nilsson D, Wincent J, et al. Replicative and non-replicative mechanisms in the formation of clustered CNVs are indicated by whole genome characterization. PLoS Genet. 2018;14:e1007780.
Pras E, Raz J, Yahalom V, Frydman M, Garzozi HJ, Pras E, et al. A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Invest Ophthalmol Vis Sci. 2004;45:1940–5.
Irum B, Khan SY, Ali M, Daud M, Kabir F, Rauf B, et al. Deletion at the GCNT2 Locus Causes Autosomal Recessive Congenital Cataracts. PloS one. 2016;11:e0167562.
Lam CW, Chan CY, Wong KC, Chang ST. Postzygotic inactivating mutation of KIF13A located at chromosome 6p22.3 in a patient with a novel mosaic neuroectodermal syndrome. J Hum Genet. 2021;66:825–9.
Donnai D, Read AP, McKeown C, Andrews T. Hypomelanosis of Ito: a manifestation of mosaicism or chimerism. J Med Genet. 1988;25:809–18.
Tang Z, Kanagal-Shamanna R, Tang G, Patel K, Medeiros LJ, Toruner GA. Analytical and clinical performance of chromosomal microarrays compared with FISH panel and conventional karyotyping in patients with chronic lymphocytic leukemia. Leuk Res. 2021;108:106616.
Mantere T, Neveling K, Pebrel-Richard C, Benoist M, van der Zande G, Kater-Baats E, et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am J Hum Genet. 2021;108:1409–22.
Alesi V, Lepri FR, Dentici ML, Genovese S, Sallicandro E, Bejo K, et al. Intragenic inversions in NF1 gene as pathogenic mechanism in neurofibromatosis type 1. Eur J Hum Genet. 2022;30:1239–43.
Chen S, Yin X, Zhang S, Xia J, Liu P, Xie P, et al. Comprehensive preimplantation genetic testing by massively parallel sequencing. Hum Reprod. 2021;36:236–47.
Hao N, Zhou J, Li M, Luo W, Zhang H, Qi Q, et al. Efficacy and initial clinical evaluation of optical genome mapping in the diagnosis of structural variations. Zhonghua Yu Fang Yi Xue. 2022;56:632–9.
Zhang S, Liang F, Lei C, Wu J, Fu J, Yang Q, et al. Long-read sequencing and haplotype linkage analysis enabled preimplantation genetic testing for patients carrying pathogenic inversions. J Med Genet. 2019;56:741–9.
Zhang S, Lei C, Wu J, Xiao M, Zhou J, Zhu S, et al. A comprehensive and universal approach for embryo testing in patients with different genetic disorders. Clin Transl Med. 2021;11:e490.
Acknowledgements
The authors appreciate the individuals and their families for their participation in this study.
Funding
The research received funding support from the National Key R&D Program of China (2021YFC1005300, 2021YFC1005301) and National High Level Hospital Clinical Research (2022-PUMCH-B-076).
Author information
Authors and Affiliations
Contributions
Conceptualization, N.H. and Y.J.; methodology, N.H. and Y.J.; software, H.L., M.L. and H.Z.; validation, M.L., H.Z., J.B., Y.W. and Y.Z.; formal analysis, N.H.; writing-original draft preparation, H.L.; writing-review and editing, N.H., J.C., Q.Q., X.Z. and Y.J.; supervision, N.H. and Y.J.; project administration, N.H. and Y.J.; funding acquisition, Y.J. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Ethics Committee of Peking Union Medical College Hospital (HS-2979). All study participants consented to undergo genetic evaluations and signed written informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hao, N., Lou, H., Li, M. et al. Analysis of complex chromosomal rearrangement involving chromosome 6 via the integration of optical genomic mapping and molecular cytogenetic methodologies. J Hum Genet 69, 3–11 (2024). https://doi.org/10.1038/s10038-023-01197-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01197-3