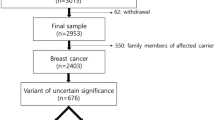
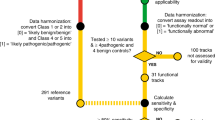
Abstract
Numerous variants of unknown significance (VUSs) exist in hereditary breast and ovarian cancers. Although multiple methods have been developed to assess the significance of BRCA1/2 variants, functional discrepancies among these approaches remain. Therefore, a comprehensive functional evaluation system for these variants should be established. We performed conventional homologous recombination (HR) assays for 50 BRCA1 and 108 BRCA2 VUSs and complementarily predicted VUSs using a statistical logistic regression prediction model that integrated six in silico functional prediction tools. BRCA1/2 VUSs were classified according to the results of the integrative in vitro and in silico analyses. Using HR assays, we identified 10 BRCA1 and 4 BRCA2 VUSs as low-functional pathogenic variants. For in silico prediction, the statistical prediction model showed high accuracy for both BRCA1 and BRCA2 compared with each in silico prediction tool individually and predicted nine BRCA1 and seven BRCA2 variants to be pathogenic. Integrative functional evaluation in this study and the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines strongly suggested that seven BRCA1 variants (p.Glu272Gly, p.Lys1095Glu, p.Val1653Leu, p.Thr1681Pro, p.Phe1761Val, p.Thr1773Ile, and p.Gly1803Ser) and four BRCA2 variants (p.Trp31Gly, p.Ser2616Phe, p.Tyr2660Cys, and p.Leu2792Arg) were pathogenic. This study demonstrates that integrative evaluation using conventional HR assays and optimized in silico prediction comprehensively classified the significance of BRCA VUSs for future clinical applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
Supplementary information is available on the Journal of Human Genetics’ website.
References
Bono M, Fanale D, Incorvaia L, Cancelliere D, Fiorino A, Calo V, et al. Impact of deleterious variants in other genes beyond BRCA1/2 detected in breast/ovarian and pancreatic cancer patients by NGS-based multi-gene panel testing: looking over the hedge. ESMO Open. 2021;6:100235.
Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207.
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71.
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92.
Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8.
Creeden JF, Nanavaty NS, Einloth KR, Gillman CE, Stanbery L, Hamouda DM, et al. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer. 2021;21:1154.
Zayas-Villanueva OA, Campos-Acevedo LD, Lugo-Trampe JJ, Hernandez-Barajas D, Gonzalez-Guerrero JF, Noriega-Iriondo MF, et al. Analysis of the pathogenic variants of BRCA1 and BRCA2 using next-generation sequencing in women with familial breast cancer: a case-control study. BMC Cancer. 2019;19:722.
Anantha RW, Simhadri S, Foo TK, Miao S, Liu J, Shen Z, et al. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. Elife. 2017;6:e21350.
Farrugia DJ, Agarwal MK, Pankratz VS, Deffenbaugh AM, Pruss D, Frye C, et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–31.
Ikegami M, Kohsaka S, Ueno T, Momozawa Y, Inoue S, Tamura K, et al. High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat Commun. 2020;11:2573.
Truty R, Ouyang K, Rojahn S, Garcia S, Colavin A, Hamlington B, et al. Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am J Hum Genet. 2021;108:696–708.
Iversen ES Jr., Lipton G, Hart SN, Lee KY, Hu C, Polley EC, et al. An integrative model for the comprehensive classification of BRCA1 and BRCA2 variants of uncertain clinical significance. NPJ Genom Med. 2022;7:35.
Guidugli L, Shimelis H, Masica DL, Pankratz VS, Lipton GB, Singh N, et al. Assessment of the clinical relevance of BRCA2 missense variants by functional and computational approaches. Am J Hum Genet. 2018;102:233–48.
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305.
Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–43.
Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32:2230–2.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7.
Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Akaike H. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G, editors. Selected Papers of Hirotugu Akaike. Springer Series in Statistics. New York: Springer; 1998. p. 199–213.
Gentleman RIR. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314.
Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5.
Momozawa Y, Iwasaki Y, Parsons MT, Kamatani Y, Takahashi A, Tamura C, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083.
Eilbeck K, Quinlan A, Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet. 2017;18:599–612.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Frazer J, Notin P, Dias M, Gomez A, Min JK, Brock K, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. 2021;599:91–5.
Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115:2222–33.
Bouwman P, van der Heijden I, van der Gulden H, de Bruijn R, Braspenning ME, Moghadasi S, et al. Functional categorization of BRCA1 variants of uncertain clinical significance in homologous recombination repair complementation assays. Clin Cancer Res. 2020;26:4559–68.
Shimelis H, Mesman RLS, Von Nicolai C, Ehlen A, Guidugli L, Martin C, et al. BRCA2 hypomorphic missense variants confer moderate risks of breast cancer. Cancer Res. 2017;77:2789–99.
Andreassen PR, Seo J, Wiek C, Hanenberg H. Understanding BRCA2 function as a tumor suppressor based on domain-specific activities in DNA damage responses. Genes (Basel). 2021;12:1034.
Poon KS. In silico analysis of BRCA1 and BRCA2 missense variants and the relevance in molecular genetic testing. Sci Rep. 2021;11:11114.
Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–38.
Thomassen M, Kruse TA, Jensen PK, Gerdes AM. A missense mutation in exon 13 in BRCA2, c.7235G>A, results in skipping of exon 13. Genet Test. 2006;10:116–20.
Fraile-Bethencourt E, Diez-Gomez B, Velasquez-Zapata V, Acedo A, Sanz DJ, Velasco EA. Functional classification of DNA variants by hybrid minigenes: Identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PLoS Genet. 2017;13:e1006691.
Acedo A, Hernandez-Moro C, Curiel-Garcia A, Diez-Gomez B, Velasco EA. Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Hum Mutat. 2015;36:210–21.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Yoshino Y, Fang Z, Qi H, Kobayashi A, Chiba N. Dysregulation of the centrosome induced by BRCA1 deficiency contributes to tissue-specific carcinogenesis. Cancer Sci. 2021;112:1679–87.
Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–63.
Mondal G, Rowley M, Guidugli L, Wu J, Pankratz VS, Couch FJ. BRCA2 localization to the midbody by filamin A regulates cep55 signaling and completion of cytokinesis. Dev Cell. 2012;23:137–52.
Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42.
Acknowledgements
JOHBOC Registration Committee [Committee]: Tadashi Nomizu (Hoshi General Hospital), Akihiro Sakurai (Sapporo Medical University), Megumi Ohkawa (St. Luke’s International Hospital), Junko Yotsumoto (International University of Health and Welfare), Kumamaru Hiraku (The University of Tokyo), [Secretariat of JOHBOC]: Shiro Yokoyama, Miyuki Shimoda. We would like to thank all the members of the Department of Oncology, Juntendo University for their valuable discussions.
Funding
This work was supported by AMED under Grant Number JP19kk0305012 (YoM), JSPS KAKENHI Grant Number JP19H03497 (YoM), JP19H01806 (MM) and JP20H04333 (SS).
Author information
Authors and Affiliations
Contributions
Conceptualization: YoM, MM, SS; Data curation: QG, SJ, KT, WU, AS, YI, HS, ZX, YuM, YoM, M.M., SS; Formal analysis: MM; Funding acquisition: YoM, MM, SS; Investigation: QG, SJ, KT, WU; Methodology: MM, SS; Project administration: YoM, MM, SS; Resources: QG, SJ, KT, WU, HS, ZX, MA, SN; Software: KT, WU, YI, MM; Supervision: YuM, NC, YoM, MM, SS; Validation: QG, SJ, KT, WU; Visualization: MM, SS; Writing-original draft: QG, SS; Writing-review & editing: YuM, YoM, MM, SS.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics Statement
Approval of the research protocol by an Institutional Reviewer Board: JOHBOC, Tokyo Medical and Dental University, and Teikyo University Ethics Committee approved the study and the use of concealed variant information from clinical databases.—Informed Consent: N/A.—Registry and the Registration No. of the study/trial: N/A.—Animal Studies: N/A.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, Q., Ji, S., Takeuchi, K. et al. Functional evaluation of BRCA1/2 variants of unknown significance with homologous recombination assay and integrative in silico prediction model. J Hum Genet 68, 849–857 (2023). https://doi.org/10.1038/s10038-023-01194-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01194-6