Abstract
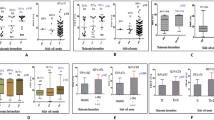
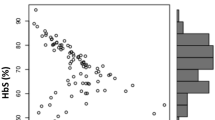
Single nucleotide polymorphisms (SNPs) of BCL11A gene and HBS1L-MYB intergenic region (named HMIP-2) affect both fetal hemoglobin (HbF) concentration and clinical outcomes in patients with sickle cell anemia (SCA). However, no previous study has examined the interaction among these SNPs in the regulation of HbF. We examined whether HbF-boosting haplotypes combining alleles of functional SNPs of BCL11A and HMIP-2 were associated with clinical outcomes and hematological parameters, and whether they interact to regulate HbF in a cohort of Brazilian children with SCA. The minor haplotype of BCL11A (“TCA”, an allele combination of rs1427407, rs766432, and rs4671393) was associated with higher HbF, hemoglobin and lower reticulocytes count compared to reference haplotype “GAG”. The minor haplotype of HMIP-2 (“CGC”, an allele combination of rs9399137, rs4895441, and rs9494145) was associated with higher HbF and hemoglobin compared to reference haplotype “TAT”. Subjects carrying minor haplotypes showed reduced rate of clinical complications compared to reference haplotypes. Non-carriers of both minor haplotypes for BCL11A and HMIP-2 showed the lowest HbF concentration. Subjects carrying only the minor haplotype of BCL11A showed significantly higher HbF concentration than non-carriers of any minor haplotype, which showed no significant difference compared to subjects carrying only the minor haplotype of HMIP-2. Interestingly, subjects carrying both minor haplotypes of BCL11A (“TCA”) and HMIP-2 (“CGC”) showed significantly higher HbF levels than subjects carrying only the minor haplotype of BCL11A. Our novel findings suggest that HbF-boosting haplotypes of BCL11A and HMIP-2 can predict clinical outcomes and may interact to regulate HbF in patients with SCA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Steinberg MH. Sickle cell anemia, the first molecular disease: overview of molecular etiology. Pathophysiol, Therapeutic Approaches Sci World J. 2008;8:1295–324.
Habara A, Steinberg MH. Minireview: genetic basis of heterogeneity and severity in sickle cell disease. Exp Biol Med. 2016;241:689–96.
Bauer DE, Orkin SH. Hemoglobin switching’s surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr Opin Genet Dev. 2015;33:62–70.
Lettre G, Sankaran VG, Bezerra MAC, Araújo AS, Uda M, Sanna S, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA. 2008;105:11869–74.
Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39:1197–99.
Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci. 2007;104:11346–51.
Bhatnagar P, Purvis S, Barron-Casella E, DeBaun MR, Casella JF, Arking DE, et al. Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J Hum Genet. 2011;56:316–23.
Sales RR, Belisario AR, Faria G, Mendes F, Luizon MR, Viana MB. Functional polymorphisms of BCL11A and HBS1L-MYB genes affect both fetal hemoglobin level and clinical outcomes in a cohort of children with sickle cell anemia. Ann Hematol. 2020;99:1453–63.
Sedgewick AE, Timofeev N, Sebastiani P, So JCC, Ma ESK, Chan LC, et al. BCL11A is a major HbF quantitative trait locus in three different populations with beta-hemoglobinopathies. Blood Cells, Molecules Dis. 2008;41:255–58.
Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105:1620–25.
Sales RR, Nogueira BL, Tosatti JAG, Gomes KB, Luizon MR. Do genetic polymorphisms affect fetal hemoglobin (HbF) levels in patients with sickle cell anemia treated with hydroxyurea? a systematic review and pathway analysis. Front in Pharmacol. 2022;12:779497.
Crawford DC, Nickerson DA. Definition and clinical importance of haplotypes. Annu Rev Med. 2005;56:303–20.
Clark AG. The role of haplotypes in candidate gene studies. Genet Epidemiol. 2004;27:321–33.
Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23:221–33.
Schaid DJ. Evaluating associations of haplotypes with traits. Genet Epidemiol. 2004;27:348–64.
Okumura JV, Silva DGH, Torres LS, Belini-Junior E, Venancio LPR, Carrocini GCS, et al. Atypical beta-S haplotypes: classification and genetic modulation in patients with sickle cell anemia. J Hum Genet. 2019;64:239–48.
Ballas SK, Lieff S, Benjamin LJ, Dampier CD, Heeney MM, Hoppe C, et al. Definitions of the phenotypic manifestations of sickle cell disease. Am J Hematol. 2010;85:6–13.
Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–59.
Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–97.
Luizon MR, Belo VA, Palei AC, Amaral LM, Lacchini R, Sandrim VC, et al. Effects of NAMPT polymorphisms and haplotypes on circulating visfatin/NAMPT levels in hypertensive disorders of pregnancy. Hypertension Research: Official. J Jpn Soc Hypertension. 2015;38:361–66.
Metzger IF, Luizon MR, Lacchini R, Ishizawa MH, Tanus-Santos JE. Effects of endothelial nitric oxide synthase tagSNPs haplotypes on nitrite levels in black subjects. Nitric Oxide. 2013;28:33–8.
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinforma (Oxf, Engl). 2005;21:263–65.
Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013;342:253–57.
Farrell JJ, Sherva RM, Chen Z-Y, Luo H-Y, Chu BF, Ha SY, et al. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood 2011;117:4935–45.
Menzel S, Thein SL. Genetic modifiers of fetal haemoglobin in sickle cell disease. Mol Diagn Ther. 2019;23:235–44.
Lettre G, Bauer DE. Fetal haemoglobin in sickle-cell disease: from genetic epidemiology to new therapeutic strategies. Lancet. 2016;387:2554–64.
Steinberg MH. Predicting clinical severity in sickle cell anaemia. Br J Haematol. 2005;129:465–81.
Martyn GE, Wienert B, Yang L, Shah M, Norton LJ, Burdach J, et al. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat Genet. 2018;50:498–503.
Fernandes APPC, Januário JN, Cangussu CB, Macedo DLD, Viana MB. Mortality of children with sickle cell disease: a population study. J Pediatr. 2010;86:279–84.
Kaul DK, Fabry ME, Nagel RL. The pathophysiology of vascular obstruction in the sickle syndromes. Blood Rev. 1996;10:29–44.
Ballas SK, Lusardi M. Hospital readmission for adult acute sickle cell painful episodes: frequency, etiology, and prognostic significance. Am J Hematol. 2005;79:17–25.
Bonds DR. Three decades of innovation in the management of sickle cell disease: the road to understanding the sickle cell disease clinical phenotype. Blood Rev. 2005;19:99–110.
Williams TN, Thein SL. Sickle cell anemia and its phenotypes. Annu Rev Genomics Hum Genet. 2018;19:113–47.
Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, et al. Pain in sickle cell disease. N Engl J Med. 1991;325:11–16.
Darbari DS, Sheehan VA, Ballas SK. The vaso-occlusive pain crisis in sickle cell disease: definition, pathophysiology, and management. Eur J Haematol. 2020;105:237–46.
Belisario AR, Sales RR, Toledo NE, Muniz MB, Velloso-Rodrigues C, Silva CM, et al. Reticulocyte count is the most important predictor of acute cerebral ischemia and high-risk transcranial Doppler in a newborn cohort of 395 children with sickle cell anemia. Ann Hematol. 2016;95:1869–80.
Leonardo FC, Brugnerotto AF, Domingos IF, Fertrin KY, de Albuquerque DM, Bezerra MA, et al. Reduced rate of sickle-related complications in Brazilian patients carrying HbF-promoting alleles at the BCL11A and HMIP-2 loci. Br J Haematol. 2016;173:456–60.
Gueye Tall F, Martin C, Ndour EHM, Renoux C, Ly ID, Connes P, et al. Combined and differential effects of alpha-thalassemia and HbF-quantitative trait loci in Senegalese hydroxyurea-free children with sickle cell anemia. Pediatr Blood Cancer. 2019;66:e27934.
Gardner K, Fulford T, Silver N, Rooks H, Angelis N, Allman M, et al. g(HbF): a genetic model of fetal hemoglobin in sickle cell disease. Blood Adv. 2018;2:235–39.
Menzel S, Rooks H, Zelenika D, Mtatiro SN, Gnanakulasekaran A, Drasar E, et al. Global genetic architecture of an erythroid quantitative trait locus, HMIP-2. Ann Hum Genet. 2014;78:434–51.
Funding
This study was supported by the Fundação Hemominas, Núcleo de Ações e Pesquisa em Apoio Diagnóstico (NUPAD/UFMG), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG/Brazil; scholarships), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil; grant 312599/2019–6), and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/Brazil; Finance Code 001).
Author information
Authors and Affiliations
Contributions
RRS, BLN, ARB, GF, FM, MBV, and MRL made substantial contributions to the conception or design of the work, acquired data, and all authors analyzed and interpreted the results. RRS, BLN, ARB, MBV, and MRL drafted the manuscript. and all authors revised it for important intellectual content. All authors read and approved the final version of the manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declared no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sales, R.R., Nogueira, B.L., Belisário, A.R. et al. Fetal hemoglobin-boosting haplotypes of BCL11A gene and HBS1L-MYB intergenic region in the prediction of clinical and hematological outcomes in a cohort of children with sickle cell anemia. J Hum Genet 67, 701–709 (2022). https://doi.org/10.1038/s10038-022-01079-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01079-0
This article is cited by
-
Effect of haplotypes of functional SNPs related to expression of fetal hemoglobin (HbF) on clinical prognosis of hematological malignancies
International Journal of Hematology (2023)