Abstract
Background
Recent researches on Parkinson’s disease (PD) pathogenesis discovered the correlation between PD and peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) dysfunction and reduction of PPARGC1A gene expression. Hence, we detected PPARGC1A rare variants to clarify their effect on PD risk in a large population of PD patients in mainland China.
Methods
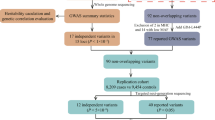
We applied whole-exome sequencing (WES) to 1917 patients with early-onset or familial PD and 1652 controls (WES cohort), and whole-genome sequencing (WGS) to 1962 patients with sporadic late-onset PD and 1279 controls (WGS cohort). To identify PPARGC1A rare variants, we used burden analysis to assess the relationship between PPARGC1A rare variants and PD susceptibility.
Results
30 rare missense variants in the cohort WES and 21 missense variants in the cohort WGS have been detected in the study and PPARGC1A missense variants are significantly associated with early-onset and familial PD susceptibility in our study (P = 0.012), which supports evidence that PPARGC1A rare variants are involved in the onset of early-onset and familial PD.
Conclusions
The study suggested that PPARGC1A rare variants may contribute to the risk of early-onset and familial PD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Schapira AHV, Chaudhuri KR, Jenner P. Non-motor features of Parkinson disease. Nat Rev Neurosci. 2017;18:435–50.
Piccinin E, Sardanelli AM, Seibel P, Moschetta A, Cocco T, Villani G. PGC-1s in the spotlight with Parkinson’s disease. Int J Mol Sci. 2021;22:3487.
Jankovic J, Tan EK. Parkinson’s disease: etiopathogenesis and treatment. J Neurol Neurosurg Psychiatry. 2020;91:795–808.
Homayoun H. Parkinson disease. Ann Intern Med. 2018;169:Itc33-itc48.
Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020;19:170–8.
Tolosa E, Garrido A, Scholz SW, Poewe W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021;20:385–97.
Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912.
Martínez-Redondo V, Pettersson AT, Ruas JL. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia. 2015;58:1969–77.
Vernier M, Giguère V. Aging, senescence and mitochondria: the PGC-1/ERR axis. J Mol Endocrinol. 2021;66:R1–14.
Nitz I, Ewert A, Klapper M, Döring F. Analysis of PGC-1α variants Gly482Ser and Thr612Met concerning their PPARγ2-coactivation function. Biochem Biophys Res Commun. 2007;353:481–6.
Jiang H, Kang S-U, Zhang S, Karuppagounder S, Xu J, Lee Y-K, et al. Adult conditional knockout of PGC-1α leads to loss of dopamine neurons. eNeuro. 2016;3:ENEURO.0183-16.2016.
Ciron C, Lengacher S, Dusonchet J, Aebischer P, Schneider BL. Sustained expression of PGC-1α in the rat nigrostriatal system selectively impairs dopaminergic function. Hum Mol Genet. 2012;21:1861–76.
Peng K, Yang L, Wang J, Ye F, Dan G, Zhao Y, et al. The interaction of mitochondrial biogenesis and fission/fusion mediated by PGC-1α regulates rotenone-induced dopaminergic neurotoxicity. Mol Neurobiol. 2017;54:3783–97.
Martínez-Redondo V, Jannig PR, Correia JC, Ferreira DMS, Cervenka I, Lindvall JM, et al. Peroxisome proliferator-activated receptor γ coactivator-1 α isoforms selectively regulate multiple splicing events on target genes. J Biol Chem. 2016;291:15169–84.
Eschbach J, von Einem B, Müller K, Bayer H, Scheffold A, Morrison BE, et al. Mutual exacerbation of peroxisome proliferator-activated receptor γ coactivator 1α deregulation and α-synuclein oligomerization. Ann Neurol. 2015;77:15–32.
Soyal SM, Zara G, Ferger B, Felder TK, Kwik M, Nofziger C, et al. The PPARGC1A locus and CNS-specific PGC-1α isoforms are associated with Parkinson’s Disease. Neurobiol Dis. 2019;121:34–46.
Yang XD, Qian YW, Xu SQ, Wan DY, Sun FH, Chen SD, et al. Expression of the gene coading for PGC-1α in peripheral blood leukocytes and related gene variants in patients with Parkinson’s disease. Parkinsonism Relat Disord. 2018;51:30–5.
Paul KC, Sinsheimer JS, Cockburn M, Bronstein JM, Bordelon Y, Ritz B. NFE2L2, PPARGC1α, and pesticides and Parkinson’s disease risk and progression. Mechanisms Ageing Dev. 2018;173:1–8.
Clark J, Reddy S, Zheng K, Betensky RA, Simon DK. Association of PGC-1alpha polymorphisms with age of onset and risk of Parkinson’s disease. BMC Med Genet. 2011;12:69–69.
Yang X, Xu S, Qian Y, He X, Chen S, Xiao Q. Hypermethylation of the gene coding for PGC-1α in peripheral blood leukocytes of patients with Parkinson’s disease. Front Neurosci. 2020;14:97.
Shi CH, Cheng Y, Tang MB, Liu YT, Yang ZH, Li F, et al. Analysis of single nucleotide polymorphisms of STK32B, PPARGC1A and CTNNA3 gene with sporadic parkinson’s disease susceptibility in Chinese Han population. Front Neurol. 2018;9:387.
Yang X, Xu S, Qian Y, He X, Chen S, Xiao Q. Hypermethylation of the gene coding for PGC-1α in peripheral blood leukocytes of patients with Parkinson’s disease. Front Neurosci. 2020;14:97.
Zhang Y, Zhao Y, Zhou X, Yi M, Li K, Zhou X, et al. Relationship between GWAS-linked three new loci in Essential tremor and risk of Parkinson’s disease in Chinese population. Parkinsonism Relat Disord. 2017;43:124–6.
Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2015;30:1591–601.
Guo JF, Zhang L, Li K, Mei JP, Xue J, Chen J, et al. Coding mutations in NUS1 contribute to Parkinson’s disease. Proc Natl Acad Sci USA. 2018;115:11567–72.
Liu H, Wang Y, Pan H, Xu K, Jiang L, Zhao Y, et al. Association of rare heterozygous PLA2G6 variants with the risk of Parkinson’s disease. Neurobiol Aging. 2021;101:297.e5–97.e8.
Pan HX, Zhao YW, Mei JP, Fang ZH, Wang Y, Zhou X, et al. GCH1 variants contribute to the risk and earlier age-at-onset of Parkinson’s disease: a two-cohort case-control study. Transl Neurodegener. 2020;9:31.
Zhao YW, Pan HX, Liu Z, Wang Y, Zeng Q, Fang ZH, et al. The association between lysosomal storage disorder genes and Parkinson’s disease: a large cohort study in Chinese mainland population. Front Aging Neurosci. 2021;13:749109.
Zhao Y, Qin L, Pan H, Liu Z, Jiang L, He Y, et al. The role of genetics in Parkinson’s disease: a large cohort study in Chinese mainland population. Brain 2020;143:2220–34.
Li J, Zhao T, Zhang Y, Zhang K, Shi L, Chen Y, et al. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46:7793–804.
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–85.
Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genom. 2013;14(Suppl 3):S3.
Huang X, Zhao Y, Pan H, Wang Y, Liu Z, Xu Q, et al. The association between LIN28A gene rare variants and Parkinson’s disease in Chinese population. Gene. 2022;829:146515.
Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–6.
Billingsley KJ, Bandres-Ciga S, Saez-Atienzar S, Singleton AB. Genetic risk factors in Parkinson’s disease. Cell Tissue Res. 2018;373:9–20.
Gureev AP, Popov VN. Nrf2/ARE pathway as a therapeutic target for the treatment of Parkinson diseases. Neurochem Res. 2019;44:2273–9.
Shi C-H, Cheng Y, Tang M-B, Liu Y-T, Yang Z-H, Li F, et al. Analysis of single nucleotide polymorphisms of STK32B, PPARGC1A and CTNNA3 gene with sporadic parkinson’s disease susceptibility in Chinese Han population. Front Neurol. 2018;9:387–7.
Müller SH, Girard SL, Hopfner F, Merner ND, Bourassa CV, Lorenz D, et al. Genome-wide association study in essential tremor identifies three new loci. Brain 2016;139:3163–9.
Jo A, Lee Y, Kam TI, Kang SU, Neifert S, Karuppagounder SS, et al. PARIS farnesylation prevents neurodegeneration in models of Parkinson’s disease. Sci Transl Med. 2021;13.
Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73.
Chen Y, Jiang Y, Yang Y, Huang X, Sun C. SIRT1 Protects Dopaminergic Neurons in Parkinson’s Disease Models via PGC-1α-Mediated Mitochondrial Biogenesis. Neurotox Res. 2021;39:1393–404.
Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 2011;144:689–702.
Acknowledgements
We thank all patients and healthy individuals for permitting us to publish their information. This study was supported by the National key R&D projects (Grant No. 2021YFC2501204) and Key Project of Hunan Provincial Science and Technology Department (Grant No. 2021SK1011) to JG, Hunan Province Innovative Construction Project (No.2019SK2335) and Science and Technology Major Project of Hunan Provincial Science and Technology Department (Grant No. 2021SK1010) to JG, the Central Public-Interest Scientific Institution Basal Research Fund of Chinese Academy of Medical Sciences (Grant No.2018-12M-HL-025) to JG, the National Natural Science Foundation of China (Grant Nos. 81873785; 81974202; 82071439; U20A20355; 82001359) to JG and BT, and the innovative team program from Department of Science & Technology of Hunan Province (Grant No. 2019RS1010, No. 2020RC4043) to JG, and the Innovation-driven Team Project from Central South University (Grant No. 2020CX016) to JG, and the National key R&D projects (Grant No. 2021YFC2502100) to JL. We are also grateful for resources from the High-Performance Computing Center of Central South University.
Author information
Authors and Affiliations
Contributions
L-ZL: Conceptualization, Methodology, Writing - original draft. YZ: Data curation, Formal analysis, Methodology, Writing - review & editing. HP: Data curation, Writing - review & editing. YX: Data curation, Writing - review & editing. YW: Data curation, Writing - review & editing. QX: Data curation, Funding acquisition. XY: Data curation, Funding acquisition. JT: Data curation, Funding acquisition. JL: Funding acquisition, Writing - review & editing. BT: Project administration, Supervision, Funding acquisition, Writing - review & editing. JG: Project administration, Supervision, Funding acquisition, Writing - review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
The study was approved by the Ethics Committee of Xiangya Hospital, Central South University and followed the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Lz., Zhao, Yw., Pan, Hx. et al. Association of rare PPARGC1A variants with Parkinson’s disease risk. J Hum Genet 67, 687–690 (2022). https://doi.org/10.1038/s10038-022-01074-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01074-5
This article is cited by
-
Current trends in basic research on Parkinson’s disease: from mitochondria, lysosome to α-synuclein
Journal of Neural Transmission (2024)