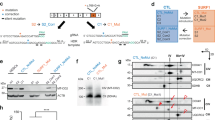
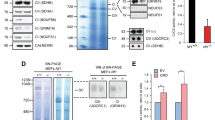
Abstract
Neurofibromatosis type 1 (NF1), one of the most common autosomal dominant genetic disorders, is caused by mutations in the NF1 gene. NF1 patients have a wide variety of manifestations with a subset at high risk for the development of tumors in the central nervous system (CNS). Nonsense mutations that result in the synthesis of truncated NF1 protein (neurofibromin) are strongly associated with CNS tumors. Therapeutic nonsense suppression with small molecule drugs is a potentially powerful approach to restore the expression of genes harboring nonsense mutations. Ataluren is one such drug that has been shown to restore full-length functional protein in several models of nonsense mutation diseases, as well as in patients with nonsense mutation Duchenne muscular dystrophy. To test ataluren’s potential applicability to NF1 nonsense mutations associated with CNS tumors, we generated a homozygous Nf1R683X/R683X-3X-FLAG mouse embryonic stem (mES) cell line which recapitulates an NF1 patient nonsense mutation (c.2041 C > T; p.Arg681X). We differentiated Nf1R683X/R683X-3X-FLAG mES cells into cortical neurons in vitro, treated the cells with ataluren, and demonstrated that ataluren can promote readthrough of the nonsense mutation at codon 683 of Nf1 mRNA in neural cells. The resulting full-length protein is able to reduce the cellular level of hyperactive phosphorylated ERK (pERK), a RAS effector normally suppressed by the NF1 protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Friedman JM. Epidemiology of neurofibromatosis type 1. Am J Med Genet. 1999;89:1–6.
Cimino PJ, Gutmann DH Chapter 51 - Neurofibromatosis type 1. In: Geschwind DH, Paulson HL, Klein C, editors. Handbook of Clinical Neurology. 148: Elsevier; 2018. p. 799-811.
Leier A, Bedwell DM, Chen AT, Dickson G, Keeling KM, Kesterson RA, et al. Mutation-Directed Therapeutics for Neurofibromatosis Type I. Mol Ther - Nucleic Acids. 2020;20:739–53.
DeClue JE, Cohen BD, Lowy DR. Identification and characterization of the neurofibromatosis type 1 protein product. Proc Natl Acad Sci. 1991;88:9914–8.
Gutmann DH, Wood DL, Collins FS. Identification of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci. 1991;88:9658–62.
Bernards A, Snijders AJ, Hannigan GE, Murthy AE, Gusella JF. Mouse neurofibromatosis type 1 cDNA sequence reveals high degree of conservation of both coding and non-coding mRNA segments. Hum Mol Genet. 1993;2:645–50.
Gutmann DH, Collins FS. The neurofibromatosis type 1 gene and its protein product, neurofibromin. Neuron 1993;10:335–43.
Xu GF, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 1990;62:599–608.
Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 1990;63:843–9.
Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, et al. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 1990;63:851–9.
Kim HA, Ratner N, Roberts TM, Stiles CD. Schwann cell proliferative responses to cAMP and Nf1 are mediated by cyclin D1. J Neurosci. 2001;21:1110–6.
Dasgupta B, Dugan LL, Gutmann DH. The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J Neurosci. 2003;23:8949–54.
Hegedus B, Dasgupta B, Shin JE, Emnett RJ, Hart-Mahon EK, Elghazi L, et al. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1:443–57.
Brown JA, Diggs-Andrews KA, Gianino SM, Gutmann DH. Neurofibromatosis-1 heterozygosity impairs CNS neuronal morphology in a cAMP/PKA/ROCK-dependent manner. Mol Cell Neurosci. 2012;49:13–22.
Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13.
Gutmann DH, Ferner RE, Listernick RH, Korf BR, Wolters PL, Johnson KJ. Neurofibromatosis type 1. Nat Rev Dis Prim. 2017;3:17004.
Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, et al. The Human Gene Mutation Database (HGMD((R))): optimizing its use in a clinical diagnostic or research setting. Hum Genet. 2020;139:1197–207.
He F, Jacobson A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu Rev Genet. 2015;49:339–66.
Osborn MJ, Upadhyaya M. Evaluation of the protein truncation test and mutation detection in the NF1 gene: mutational analysis of 15 known and 40 unknown mutations. Hum Genet. 1999;105:327–32.
Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat Res. 2014;762:52–64.
Bashyam MD. Nonsense-mediated decay: linking a basic cellular process to human disease. Expert Rev Mol Diagn. 2009;9:299–303.
Anastasaki C, Morris SM, Gao F, Gutmann DH. Children with 5’-end NF1 gene mutations are more likely to have glioma. Neurol Genet. 2017;3:e192.
Brinckmann A, Mischung C, Bassmann I, Kuhnisch J, Schuelke M, Tinschert S, et al. Detection of novel NF1 mutations and rapid mutation prescreening with Pyrosequencing. Electrophoresis 2007;28:4295–301.
Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics Based on Stop Codon Readthrough. Annu Rev Genomics Hum Genet. 2014;15:8.1–8.
Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–25.
Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91.
Li M, Andersson-Lendahl M, Sejersen T, Arner A. Muscle dysfunction and structural defects of dystrophin-null sapje mutant zebrafish larvae are rescued by ataluren treatment. FASEB J. 2014;28:1593–9.
Gregory-Evans CY, Wang X, Wasan KM, Zhao J, Metcalfe AL, Gregory-Evans K. Postnatal manipulation of Pax6 dosage reverses congenital tissue malformation defects. J Clin Invest. 2014;124:111–6.
Bushby K, Finkel R, Wong B, Barohn R, Campbell C, Comi GP, et al. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014;50:477–87.
Ryan NJ. Ataluren: first global approval. Drugs 2014;74:1709–14.
Li K, Turner AN, Chen M, Brosius SN, Schoeb TR, Messiaen LM, et al. Mice with missense and nonsense NF1 mutations display divergent phenotypes compared with human neurofibromatosis type I. Dis Model Mech. 2016;9:759–67.
Kocova M, Kochova E, Sukarova-Angelovska E. Optic glioma and precocious puberty in a girl with neurofibromatosis type 1 carrying an R681X mutation of NF1: case report and review of the literature. BMC Endocr Disord. 2015;15:82.
Toonen JA, Anastasaki C, Smithson LJ, Gianino SM, Li K, Kesterson RA, et al. NF1 germline mutation differentially dictates optic glioma formation and growth in neurofibromatosis-1. Hum Mol Genet. 2016;25:1703–13.
Gaspard N, Bouschet T, Herpoel A, Naeije G, van den Ameele J, Vanderhaeghen P. Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc. 2009;4:1454–63.
Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature. 2019;568:561–5.
Bae S, Park J, Kim J-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014;30:1473–5.
Costa ADA, Gutmann DH Brain tumors in neurofibromatosis type 1. Neuro-Oncology Advances. 2019;2(Supplement_1):i85-i97.
Guilding C, McNair K, Stone TW, Morris BJ. Restored plasticity in a mouse model of neurofibromatosis type 1 via inhibition of hyperactive ERK and CREB. Eur J Neurosci. 2007;25:99–105.
Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, et al. Neurofibromin Regulation of ERK Signaling Modulates GABA Release and Learning. Cell 2008;135:549–60.
Pan Y, Hysinger JD, Barron T, Schindler NF, Cobb O, Guo X, et al. NF1 mutation drives neuronal activity-dependent initiation of optic glioma. Nature. 2021;594:277–82.
Ars E, Kruyer H, Morell M, Pros E, Serra E, Ravella A, et al. Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet. 2003;40:e82.
Ben-Salem S, Al-Shamsi AM, Ali BR, Al-Gazali L. The mutational spectrum of the NF1 gene in neurofibromatosis type I patients from UAE. Childs Nerv Syst. 2014;30:1183–9.
N Abdel-Aziz N, Y El-Kamah G, A Khairat R, R Mohamed H, Z Gad Y, El-Ghor AM, et al. Mutational spectrum of NF1 gene in 24 unrelated Egyptian families with neurofibromatosis type 1. Mol Genet Genom Med. 2021;9:e1631.
Ko JM, Sohn YB, Jeong SY, Kim HJ, Messiaen LM. Mutation spectrum of NF1 and clinical characteristics in 78 Korean patients with neurofibromatosis type 1. Pediatr Neurol. 2013;48:447–53.
Acknowledgements
This work was supported by grants to AJ (616394) and SAW (563749) from the Gilbert Family Foundation. We thank Pengpeng Liu and Sneha Suresh for helpful discussions, and PTC Therapeutics Inc. for providing ataluren.
Author information
Authors and Affiliations
Contributions
CW and AJ conceived and designed the work. CW performed experiments. SI and SAW provided technical support. CW and AJ wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
AJ is co-founder, director, and SAB chair of PTC Therapeutics Inc. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, C., Iyer, S., Wolfe, S.A. et al. Functional restoration of mouse Nf1 nonsense alleles in differentiated cultured neurons. J Hum Genet 67, 661–668 (2022). https://doi.org/10.1038/s10038-022-01072-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01072-7