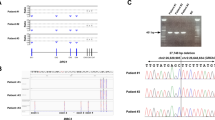
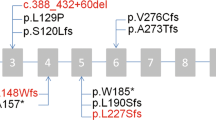
Abstract
Primary ciliary dyskinesia (PCD) is a clinically and genetically heterogeneous ciliopathy. Dysfunction of motile respiratory and nodal cilia results in sinopulmonary symptoms associated with laterality defects (LD) found in half of the patients. The molecular basis of the disease is insufficiently investigated in patients originating from the Arabian Peninsula. In a group of 16 unrelated Saudi patients clinically suspected of PCD and among whom only 5 (31%) had LD, we first screened by PCR-RFLP two founder mutations, RSPH9 c.804_806del and CCDC39 c.2190del previously identified in patients from the Arabian Peninsula and Tunisia, respectively. When negative, targeted panel or whole-exome sequencing was performed. Three patients were homozygous for the mutation in RSPH9, which encodes an axonemal protein that is absent from nodal cilia. None of the patients carried the CCDC39 founder mutation frequent in Tunisia. NGS analysis showed that nine patients had homozygous mutations in PCD genes. In total, sequential RFLP and NGS analysis solved 75% (12/16) of cases and identified ten distinct mutations, among which six are novel, in nine different genes. These results, which highlight the genetic heterogeneity of PCD in Saudi Arabia, show that the RSPH9 c.804_806del mutation is a prevalent mutation among Saudi patients, whereas the CCDC39 c.2190del ancestral allele is most likely related to the Berber population. This study shows that RSPH9 founder mutation first-line screening and NGS analysis is efficient for the genetic exploration of PCD in Saudi patients. The RSPH9 founder mutation accounts for the low rate of LD among Saudi patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–9.
Afzelius BA, Srurgess JM. The immotile-cilia syndrome: a microtubule-associated defect. Crit Rev Biochem. 1985;19:63–87.
Mullowney T, Manson D, Kim R, Stephens D, Shah V, Dell S. Primary ciliary dyskinesia and neonatal respiratory distress. Pediatrics. 2014;134:1160–6.
Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999;4:459–68.
Leigh MW, Ferkol TW, Davis SD, Lee HS, Rosenfeld M, Dell SD, et al. Clinical features and associated likelihood of primary ciliary dyskinesia in children and adolescents. Ann Am Thorac Soc. 2016;13:1305–13.
Lucas JS, Barbato A, Collins SA, Goutaki M, Behan L, Caudri D, et al. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Respir J. 2017;49:1601090.
Shapiro AJ, Leigh MW. Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: genetic defects with normal and non-diagnostic ciliary ultrastructure. Ultrastruct Pathol. 2017;41:373–85.
Shoemark A, Frost E, Dixon M, Ollosson S, Kilpin K, Patel M, et al. Accuracy of immunofluorescence in the diagnosis of primary ciliary dyskinesia. Am J Respir Crit Care Med. 2017;196:94–101.
Rubbo B, Shoemark A, Jackson CL, Hirst R, Thompson J, Hayes J, et al. Accuracy of high-speed video analysis to diagnose primary ciliary dyskinesia. Chest. 2019;155:1008–17.
Legendre M, Zaragosi L-E, Mitchison HM. Motile cilia and airway disease. Semin Cell Dev Biol. 2021;110:19–33.
Wallmeier J, Frank D, Shoemark A, Nöthe-Menchen T, Cindric S, Olbrich H, et al. De novo mutations in FOXJ1 result in a motile ciliopathy with hydrocephalus and randomization of left/right body asymmetry. Am J Hum Genet. 2019;105:1030–9.
Olcese C, Patel MP, Shoemark A, Kiviluoto S, Legendre M, Williams HJ, et al. X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat Commun. 2017;8:14279.
Hannah WB, DeBrosse S, Kinghorn B, Strausbaugh S, Aitken ML, Rosenfeld M, et al. The expanding phenotype of OFD1‐related disorders: hemizygous loss‐of‐function variants in three patients with primary ciliary dyskinesia. Mol Genet Genomic Med. 2019;7:e911. https://doi.org/10.1002/mgg3.911.
Moore A, Escudier E, Roger G, Tamalet A, Pelosse B, Marlin S, et al. RPGR is mutated in patients with a complex X linked phenotype combining primary ciliary dyskinesia and retinitis pigmentosa. J Med Genet. 2006;43:326–33.
Paff T, Loges NT, Aprea I, Wu K, Bakey Z, Haarman EG, et al. Mutations in PIH1D3 cause X-linked primary ciliary dyskinesia with outer and inner dynein arm defects. Am J Hum Genet. 2017;100:160–8.
Zariwala MA, Knowles MR, Leigh MW. Primary ciliary dyskinesia. 5 December 2019. University of Washington, Seattle. 2019. http://www.ncbi.nlm.nih.gov/pubmed/20301301. Accessed 18 July 2021.
Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, Van Arendonk LG, et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2012;91:685–93.
Knowles MR, Ostrowski LE, Loges NT, Hurd T, Leigh MW, Huang L, et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am J Hum Genet. 2013;93:711–20.
Boaretto F, Snijders D, Salvoro C, Spalletta A, Mostacciuolo ML, Collura M, et al. Diagnosis of primary ciliary dyskinesia by a targeted next-generation sequencing panel: molecular and clinical findings in Italian patients. J Mol Diagnostics. 2016;18:912–22.
Mani R, Belkacem S, Soua Z, Chantot S, Montantin G, Tissier S, et al. Primary ciliary dyskinesia gene contribution in Tunisia: identification of a major Mediterranean allele. Hum Mutat. 2020;41:115–21.
Fassad MR, Shoman WI, Morsy H, Patel MP, Radwan N, Jenkins L, et al. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin Genet. 2020;97:509–15.
Horani A, Ferkol TW. Advances in the genetics of primary ciliary dyskinesia. Chest. 2018;154:645–52.
Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia: recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–22.
Knowles MR, Leigh MW. Primary ciliary dyskinesia diagnosis. is color better than black and white? Am J Respir Crit Care Med. 2017;196:9–10.
Zariwala MA, Leigh MW, Ceppa F, Kennedy MP, Noone PG, Carson JL, et al. Mutations of DNAI1 in primary ciliary dyskinesia evidence of founder effect in a common mutation. Am J Respir Crit Care Med. 2006;174:858–66.
Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges NT, Raidt J, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91:672–84.
AlSaadi MM, Gaunt TR, Boustred CR, Guthrie PA, Liu X, Lenzi L, et al. From a single whole exome read to notions of clinical screening: primary ciliary dyskinesia and RSPH9 p.Lys268del in the Arabian Peninsula. Ann Hum Genet. 2012;76:211–20.
Shamseldin HE, Al Mogarri I, Alqwaiee MM, Alharbi AS, Baqais K, AlSaadi M, et al. An exome-first approach to aid in the diagnosis of primary ciliary dyskinesia. Hum Genet. 2020;139:1273–83.
Mani R, Belkacem S, Soua Z, Chantot S, Montantin G, Tissier S, et al. Primary ciliary dyskinesia gene contribution in Tunisia—identification of a major Mediterranean allele. Hum Mutat. 2020;41:115–121.
Dimassi S, Simonet T, Labalme A, Boutry-Kryza N, Campan-Fournier A, Lamy R, et al. Comparison of two next-generation sequencing kits for diagnosis of epileptic disorders with a user-friendly tool for displaying gene coverage, DeCovA. Appl Transl Genomics. 2015;7:19–25.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24.
Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Marshall CR, Scherer SW, Zariwala MA, Lau L, Paton TA, Stockley T, et al. Whole-exome sequencing and targeted copy number analysis in primary ciliary dyskinesia. G3 Genes|Genomes|Genet. 2015;5:1775–81.
Djakow J, Kramná L, Dušátková L, Uhlík J, Pursiheimo JP, Svobodová T, et al. An effective combination of sanger and next generation sequencing in diagnostics of primary ciliary dyskinesia. Pediatr Pulmonol. 2016;509:498–509.
Baz-Redón N, Rovira-Amigo S, Paramonov I, Castillo-Corullón S, Cols Roig M, Antolín M, et al. Implementación de un panel de genes para el diagnóstico genético de la discinesia ciliar primaria. Arch Bronconeumol. 2020;57:186–94. https://doi.org/10.1016/j.arbres.2020.02.010.
Emiralioğlu N, Taşkıran EZ, Koşukcu C, Bilgiç E, Atilla P, Kaya B, et al. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: first results from Turkey. Pediatr Pulmonol. 2020;55:383–93.
Chivukula RR, Montoro DT, Leung HM, Yang J, Shamseldin HE, Taylor MS, et al. A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance. Nat Med. 2020;26:244–51.
Al Mutairi F, Alkhalaf R, Alkhorayyef A, Alroqi F, Yusra A, Umair M, et al. Homozygous truncating NEK10 mutation, associated with primary ciliary dyskinesia: a case report. BMC Pulm Med. 2020;20:141.
Mitchison HM, Schmidts M, Loges NT, Freshour J, Dritsoula A, Hirst RA, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–9.
Erzurumluoglu AM. Population and family based studies of consanguinity: genetic and computational approaches. University of Bristol; 2015.
Reish O, Slatkin M, Chapman-Shimshoni D, Elizur A, Chioza B, Castleman V, et al. Founder mutation(s) in the RSPH9 gene leading to primary ciliary dyskinesia in two inbred Bedouin families. Ann Hum Genet. 2010;74:117–25.
Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–8.
Acknowledgements
Authors are grateful to the Chancellerie des Universités of Sorbonne Université for its support through the Legs Poix grant. French authors and data contributors participate in the BEAT-PCD clinical research collaboration, supported by the European Respiratory Society. We thank families for their participation in this study.
Funding
This research was supported by Deanship of Scientific Research, Taif University, Saudi Arabia (Research group number 1-440-6148).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mabrouk, I., Al-Harthi, N., Mani, R. et al. Combining RSPH9 founder mutation screening and next-generation sequencing analysis is efficient for primary ciliary dyskinesia diagnosis in Saudi patients. J Hum Genet 67, 381–386 (2022). https://doi.org/10.1038/s10038-021-01006-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-01006-9