Abstract
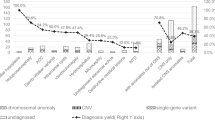
Corpus callosum anomalies (CCA) is a common congenital brain anomaly with various etiologies. Although one of the most important etiologies is genetic factors, the genetic background of CCA is heterogenous and diverse types of variants are likely to be causative. In this study, we analyzed 16 Japanese patients with corpus callosum anomalies to delineate clinical features and the genetic background of CCAs. We observed the common phenotypes accompanied by CCAs: intellectual disability (100%), motor developmental delay (93.8%), seizures (60%), and facial dysmorphisms (50%). Brain magnetic resonance imaging showed colpocephaly (enlarged posterior horn of the lateral ventricles, 84.6%) and enlarged supracerebellar cistern (41.7%). Whole exome sequencing revealed genetic alterations in 9 of the 16 patients (56.3%), including 8 de novo alterations (2 copy number variants and variants in ARID1B, CDK8, HIVEP2, and TCF4) and a recessive variant of TBCK. De novo ARID1B variants were identified in three unrelated individuals, suggesting that ARID1B variants are major genetic causes of CCAs. A de novo TCF4 variant and somatic mosaic deletion at 18q21.31-qter encompassing TCF4 suggest an association of TCF4 abnormalities with CCAs. This study, which analyzes CCA patients usung whole exome sequencing, demonstrates that comprehensive genetic analysis would be useful for investigating various causal variants of CCAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the supplementary material of this article.
References
Raybaud C. Corpus callosum: molecular pathways in mice and human dysgeneses. Neuroimaging Clin N Am. 2019;29:445–59.
van der Knaap LJ, van der Ham IJ. How does the corpus callosum mediate interhemispheric transfer? A review. Behav Brain Res. 2011;223:211–21.
van Wagenen WP, Herren RY. Surgical division of commissural pathways in the corpus callosum. Arch Neurol Psychiatry. 1940;44:740–59.
Gazzaniga MS. Forty-five years of split-brain research and still going strong. Nat Rev Neurosci. 2005;6:653–9.
Al-Hashim AH, Blaser S, Raybaud C, MacGregor D. Corpus callosum abnormalities: neuroradiological and clinical correlations. Dev Med Child Neurol. 2016;58:475–84.
Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain 2014;137:1579–613.
Sotiriadis A, Makrydimas G. Neurodevelopment after prenatal diagnosis of isolated agenesis of the corpus callosum: an integrative review. Am J Obstet Gynecol. 2012;206:337.
Schell-Apacik CC, Wagner K, Bihler M, Ertl-Wagner B, Heinrich U, Klopocki E, et al. Agenesis and dysgenesis of the corpus callosum: clinical, genetic and neuroimaging findings in a series of 41 patients. Am J Med Genet A 2008;146A:2501–11.
Bedeschi MF, Bonaglia MC, Grasso R, Pellegri A, Garghentino RR, Battaglia MA, et al. Agenesis of the corpus callosum: clinical and genetic study in 63 young patients. Pediatr Neurol. 2006;34:186–93.
Hiraide T, Nakashima M, Ikeda T, Tanaka D, Osaka H, Saitsu H. Identification of a deep intronic POLR3A variant causing inclusion of a pseudoexon derived from an Alu element in Pol III-related leukodystrophy. J Hum Genet. 2020;65:921–5.
Shiohama T, Nakashima M, Ikehara H, Kato M, Saitsu H. Low-prevalence mosaicism of chromosome 18q distal deletion identified by exome-based copy number profiling in a child with cerebral hypomyelination. Congenit Anom. 2020;60:94–6.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Chong JX, Caputo V, Phelps IG, Stella L, Worgan L, Dempsey JC, et al. Recessive inactivating mutations in TBCK, encoding a Rab GTPase-activating protein, cause severe infantile syndromic encephalopathy. Am J Hum Genet. 2016;98:772–81.
Bhoj EJ, Li D, Harr M, Edvardson S, Elpeleg O, Chisholm E, et al. Mutations in TBCK, encoding TBC1-domain-containing kinase, lead to a recognizable syndrome of intellectual disability and hypotonia. Am J Hum Genet. 2016;98:782–8.
Mignot C, Moutard ML, Rastetter A, Boutaud L, Heide S, Billette T, et al. ARID1B mutations are the major genetic cause of corpus callosum anomalies in patients with intellectual disability. Brain 2016;1:e64.
Calpena E, Hervieu A, Kaserer T, Swagemakers SMA, Goos JAC, Popoola O, et al. De novo missense substitutions in the gene encoding CDK8, a regulator of the mediator complex, cause a syndromic developmental disorder. Am J Hum Genet. 2019;4:104.
Goodspeed K, Newsom C, Morris MA, Powell C, Evans P, Golla S. Pitt-Hopkins syndrome: a review of current literature, clinical approach, and 23-patient case series. J Child Neurol. 2018;33:233–44.
Srivastava S, Engels H, Schanze I, Cremer K, Wieland T, Menzel M, et al. Loss-of-function variants in HIVEP2 are a cause of intellectual disability. Eur J Hum Genet. 2016;24:556–61.
Sajan SA, Fernandez L, Nieh SE, Rider E, Bukshpun P, Wakahiro M, et al. Both rare and de novo copy number variants are prevalent in agenesis of the corpus callosum but not in cerebellar hypoplasia or polymicrogyria. PLoS Genet. 2013;9:e1003823.
Heide S, Keren B, Billette de Villemeur T, Chantot-Bastaraud S, Depienne C, Nava C, et al. Copy number variations found in patients with a corpus callosum abnormality and intellectual disability. J Pediatr. 2017;185:160.
Liao LH, Chen C, Peng J, Wu LW, He F, Yang LF, et al. Diagnosis of intellectual disability/global developmental delay via genetic analysis in a central region of China. Chin Med J. 2019;132:1533–40.
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46.
van der Schoot V, de Munnik S, Venselaar H, Elting M, Mancini GMS, Ravenswaaij-Arts CMA, et al. Toward clinical and molecular understanding of pathogenic variants in the ZBTB18 gene. Mol Genet Genom Med. 2018;6:393–400.
Shetty M, Srikanth A, Kadandale J, Hegde S. Pre- and postnatal analysis of chromosome 1q44 deletion in agenesis of corpus callosum. Mol Syndromol. 2015;6:187–92.
Hasi M, Soileau B, Sebold C, Hill A, Hale DE, O’Donnell L, et al. The role of the TCF4 gene in the phenotype of individuals with 18q segmental deletions. Hum Genet. 2011;130:777–87.
Phan BN, Bohlen JF, Davis BA, Ye Z, Chen HY, Mayfield B, et al. A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder. Nat Neurosci. 2020;23:375–85.
Ebrahimi-Fakhari D, Saffari A, Wahlster L, Lu J, Byrne S, Hoffmann GF, et al. Congenital disorders of autophagy: an emerging novel class of inborn errors of neuro-metabolism. Brain 2016;139:317–37.
Crino PB. mTOR signaling in epilepsy: insights from malformations of cortical development. Cold Spring Harb Perspect Med. 2015;5:a022442.
Ortiz-González XR, Tintos-Hernández JA, Keller K, Li X, Foley AR, Bharucha-Goebel DX, et al. Homozygous boricua TBCK mutation causes neurodegeneration and aberrant autophagy. Ann Neurol. 2018;83:153–65.
Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy 2013;9:124–37.
Moffat JJ, Jung EM, Ka M, Smith AL, Jeon BT, Santen GWE, et al. The role of ARID1B, a BAF chromatin remodeling complex subunit, in neural development and behavior. Prog Neuropsychopharmacol Biol Psychiatry. 2019;8:89.
Lieb JM, Ahlhelm FJ. [Agenesis of the corpus callosum]. Radiologe 2018;58:636–45.
Dutta I, Sharma GN, Singh KP. Transsphenoidal encephalocele, colpocephaly and corpus callosum agenesis in a midline cleft lip and palate patient: a very rare case. Indian J Plast Surg. 2018;51:334–5.
Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene 2009;28:1653–68.
Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–8.
Filatova A, Rey LK, Lechler MB, Schaper J, Hempel M, Posmyk R, et al. Mutations in SMARCB1 and in other Coffin-Siris syndrome genes lead to various brain midline defects. Nat Commun. 2019;10:2966.
van der Sluijs PJ, Jansen S, Vergano SA, Adachi-Fukuda M, Alanay Y, AlKindy A, et al. The ARID1B spectrum in 143 patients: from nonsyndromic intellectual disability to Coffin-Siris syndrome. Genet Med. 2019;21:1295–307.
Shimbo H, Yokoi T, Aida N, Mizuno S, Suzumura H, Nagai J, et al. Haploinsufficiency of BCL11A associated with cerebellar abnormalities in 2p15p16.1 deletion syndrome. Mol Genet Genom Med. 2017;5:429–37.
Huang HT, Chen SM, Pan LB, Yao J, Ma HT. Loss of function of SWI/SNF chromatin remodeling genes leads to genome instability of human lung cancer. Oncol Rep. 2015;33:283–91.
Acknowledgements
The authors would like to thank the patient’s family for participating in this study. The authors also thank K. Shibazaki, M. Tsujimura, and A. Kitamoto for their technical assistance. This study was supported by Grant‐in‐Aid from the Ministry of Health, Labour and Welfare of Japan, the Takeda Science Foundation, and HUSM Grant-in-Aid from Hamamatsu University School of Medicine.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Miyamoto, S., Kato, M., Hiraide, T. et al. Comprehensive genetic analysis confers high diagnostic yield in 16 Japanese patients with corpus callosum anomalies. J Hum Genet 66, 1061–1068 (2021). https://doi.org/10.1038/s10038-021-00932-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00932-y