Abstract
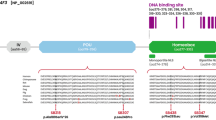
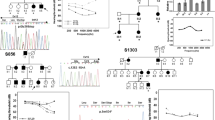
Mutations in the CDC14A (Cell Division-Cycle 14A) gene, which encodes a conserved dual-specificity protein tyrosine phosphatase, have been identified as a cause of autosomal recessive non-syndromic hearing loss (DFNB32) and hearing impairment infertility male syndrome (HIIMS). We used next-generation sequencing to screen six deaf probands from six families segregating sensorineural moderate-to-profound hearing loss. Data analysis and variant prioritization were completed using a custom bioinformatics pipeline. We identified three homozygous loss of function variants (p.Arg345Ter, p.Arg376Ter, and p.Ala451Thrfs*43) in the CDC14A gene, segregating with deafness in each family. Of the six families, four segregated the p.Arg376Ter mutation, one family segregated the p.Arg345Ter mutation and one family segregated a novel frameshift (p.Ala451Thrfs*43) mutation. In-depth phenotyping of affected individuals ruled out secondary syndromic findings. This study implicates the p.Arg376Ter mutation might be as a founder mutation in the Iranian population. It also provides the first semen analysis for deaf males carrying mutations in exon 11 of CDC14A and reveals a genotype–phenotype correlation that delineates between DFNB32 and HIIMS. The clinical results from affected males suggest the NM_033313.2 transcript alone is sufficient for proper male fertility, but not for proper auditory function. We conclude that DFNB32 is a distinct phenotypic entity in males.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in Clinvar database at https://www.ncbi.nlm.nih.gov/clinvar/docs/submit/, reference number: SUB5843570.
References
Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–89.
Uddin B, Partscht P, Chen NP, Neuner A, Weiss M, Hardt R, et al. The human phosphatase CDC14A modulates primary cilium length by regulating centrosomal actin nucleation. EMBO Rep. 2019;20:e46544.
Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, et al. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol. 2010;189:631–9.
Clement A, Solnica-Krezel L, Gould KL. The Cdc14B phosphatase contributes to ciliogenesis in zebrafish. Development. 2011;138:291–302.
Chen NP, Uddin B, Voit R, Schiebel E. Human phosphatase CDC14A is recruited to the cell leading edge to regulate cell migration and adhesion. Proc Natl Acad Sci USA. 2016;113:990–5.
Lin H, Ha K, Lu G, Fang X, Cheng R, Zuo Q, et al. Cdc14A and Cdc14B redundantly regulate DNA double-strand break repair. Mol Cell Biol. 2015;35:3657–68.
Ovejero S, Ayala P, Bueno A, Sacristan MP. Human Cdc14A regulates Wee1 stability by counteracting CDK-mediated phosphorylation. Mol Biol Cell. 2012;23:4515–25.
Bremmer SC, Hall H, Martinez JS, Eissler CL, Hinrichsen TH, Rossie S, et al. Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J Biol Chem. 2012;287:1662–9.
Vazquez-Novelle MD, Mailand N, Ovejero S, Bueno A, Sacristan MP. Human Cdc14A phosphatase modulates the G2/M transition through Cdc25A and Cdc25B. J Biol Chem. 2010;285:40544–53.
Schindler K, Schultz RM. The CDC14A phosphatase regulates oocyte maturation in mouse. Cell Cycle. 2009;8:1090–8.
Schindler K, Schultz RM. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod. 2009;80:795–803.
Trautmann S, McCollum D. Cell cycle: new functions for Cdc14 family phosphatases. Curr Biol. 2002;12:R733–5.
Bembenek J, Yu H. Regulation of the anaphase-promoting complex by the dual specificity phosphatase human Cdc14a. J Biol Chem. 2001;276:48237–42.
Delmaghani S, Aghaie A, Bouyacoub Y, El Hachmi H, Bonnet C, Riahi Z, et al. Mutations in CDC14A, encoding a protein phosphatase involved in hair cell ciliogenesis, cause autosomal-recessive severe to profound deafness. Am J Hum Genet. 2016;98:1266–70.
Imtiaz A, Belyantseva IA, Beirl AJ, Fenollar-Ferrer C, Bashir R, Bukhari I, et al. CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum Mol Genet. 2018;27:780–98.
Doll J, Kolb S, Schnapp L, Rad A, Ruschendorf F, Khan I, et al. Novel loss-of-function variants in CDC14A are associated with recessive sensorineural hearing loss in Iranian and Pakistani patients. Int J Mol Sci. 2020;21:E311.
Booth KT, Azaiez H, Kahrizi K, Wang D, Zhang Y, Frees K, et al. Exonic mutations and exon skipping: lessons learned from DFNA5. Hum Mutat. 2018;39:433–40.
Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F, et al. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet. 2015;52:823–9.
Booth KT, Azaiez H, Kahrizi K, Simpson AC, Tollefson WT, Sloan CM, et al. PDZD7 and hearing loss: More than just a modifier. Am J Med Genet A. 2015;167a:2957–65.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
The Genomes Project C, Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, et al. A global reference for human genetic variation. Nature. 2015;526:68.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 2019:531210. https://www.biorxiv.org/content/10.1101/531210v2.
Fattahi Z, Beheshtian M, Mohseni M, Poustchi H, Sellars E, Nezhadi SH, et al. Iranome: a catalog of genomic variations in the Iranian population. Hum Mutat. 2019;40:1968–84.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7.
Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13.
Gray CH, Good VM, Tonks NK, Barford D. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. Embo J. 2003;22:3524–35.
Mocciaro A, Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci. 2010;123:2867–76.
Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–9.
Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–45.
Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–97.
Lentz J, Keats B. Usher syndrome type II. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews((R)). Seattle (WA): University of Washington; 1993.
Lentz J, Keats BJB. Usher Syndrome Type I In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews((R)). Seattle (WA): University of Washington; 1993.
Booth KT, Askew JW, Talebizadeh Z, Huygen PLM, Eudy J, Kenyon J, et al. Splice-altering variant in COL11A1 as a cause of nonsyndromic hearing loss DFNA37. Genet Med. 2019;21:948–54.
Robin NH, Moran RT, Ala-Kokko L Stickler Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews((R)). Seattle (WA): University of Washington; 1993.
Zazo Seco C, Serrao de Castro L, van Nierop JW, Morin M, Jhangiani S, Verver EJ, et al. Allelic mutations of KITLG, encoding KIT ligand, cause asymmetric and unilateral hearing loss and waardenburg syndrome type 2. Am J Hum Genet. 2015;97:647–60.
Acknowledgements
We are grateful to the patients and families for their valuable collaboration. KTB was supported by NIH/NIGMS grant T32 GM007748.
Funding
This work was supported by the Iran National Science Foundation (INSF) (grant number: 96011200).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mohseni, M., Akbari, M., Booth, K.T. et al. When transcripts matter: delineating between non-syndromic hearing loss DFNB32 and hearing impairment infertile male syndrome (HIIMS). J Hum Genet 65, 609–617 (2020). https://doi.org/10.1038/s10038-020-0740-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0740-z
This article is cited by
-
Genetic etiology of hearing loss in Iran
Human Genetics (2022)