Abstract
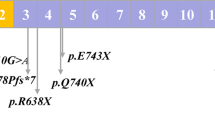
Hereditary spherocytosis (HS) is the most common inherited hemolytic anemia characterized by the presence of spherical-shaped erythrocytes on the peripheral blood smear, hemolysis, splenomegaly, jaundice, and gallstones. To date, mutations in at least five genes (ANK1, EPB42, SLC4A1, SPTA1, and SPTB) have been found to be associated with different subtypes of HS. Here, we aim to investigate the presence of novel as well as known mutations in 35 Chinese patients with clinically suspected HS. Whole-exome sequencing (WES) has identified 3 patients with SLC4A1, 16 patients with ANK1, and 16 patients with SPTB mutations, including 5 splicing, 12 nonsense, 9 frameshift, 7 missense, and 1 start-loss mutation, indicating that SPTB and ANK1 are the most frequently mutated genes in Chinese HS patients. Among 34 mutations identified, 21 were novel. Most of SPTB and ANK1 mutations were nonsense (8/16) and frameshift (6/16) mutations. By trio analysis of eight families we have confirmed six de novo mutations. In addition, genotype–phenotype analysis was also performed by comparing clinical manifestations among three groups of patients with SPTB, ANK1, and SLC4A1 mutations. It revealed that patients with ANK1 mutations had a significantly higher level of MCV and MCH but lower percentage of spherocytes compared with those carrying SPTB mutations. In conclusion, our results suggested that molecular diagnosis by next-generation sequencing (NGS) is a fast, economic, and accurate way to detect and identify pathogenic alterations of inherited diseases, highlighting the potential usage of NGS in clinical practice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Perrotta S, Gallagher PG, Mohandas N. Hereditary spherocytosis. Lancet 2008;372:1411–26.
Wang C, Cui Y, Li Y, Liu X, Han J. A systematic review of hereditary spherocytosis reported in Chinese biomedical journals from 1978 to 2013 and estimation of the prevalence of the disease using a disease model. Intractable Rare Dis Res 2015;4:76–81.
Gallagher PG. Red cell membrane disorders. Hematol Am Soc Hematol Educ Program. 2005:13–8. https://doi.org/10.1182/asheducation-2005.1.13.
Barcellini W, Bianchi P, Fermo E, Imperiali FG, Marcello AP, Vercellati C, et al. Hereditary red cell membrane defects: diagnostic and clinical aspects. Blood Transfus 2011;9:274–7.
Da Costa L, Galimand J, Fenneteau O, Mohandas N. Hereditary spherocytosis, elliptocytosis, and other red cell membrane disorders. Blood Rev. 2013;27:167–78.
Delaunay J. The molecular basis of hereditary red cell membrane disorders. Blood Rev. 2007;21:1–20.
An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367–75.
Delaunay J. Molecular basis of red cell membrane disorders. Acta Haematol. 2002;108:210–8.
Eber SW, Armbrust R, Schroter W. Variable clinical severity of hereditary spherocytosis: relation to erythrocytic spectrin concentration, osmotic fragility, and autohemolysis. J Pediatr. 1990;117:409–16.
Park J, Jeong DC, Yoo J, Jang W, Chae H, Kim J, et al. Mutational characteristics of ANK1 and SPTB genes in hereditary spherocytosis. Clin Genet. 2016;90:69–78.
Wang R, Yang S, Xu M, Huang J, Liu H, Gu W, et al. Exome sequencing confirms molecular diagnoses in 38 Chinese families with hereditary spherocytosis. Sci China Life Sci. 2018;61:947–53.
Leite RC, Basseres DS, Ferreira JS, Alberto FL, Costa FF, Saad ST. Low frequency of ankyrin mutations in hereditary spherocytosis: identification of three novel mutations. Hum Mutat. 2000;16:529.
Yawata Y, Kanzaki A, Yawata A, Doerfler W, Ozcan R, Eber SW. Characteristic features of the genotype and phenotype of hereditary spherocytosis in the Japanese population. Int J Hematol. 2000;71:118–35.
Gao Y, Zhang B, Song Y, Li G, Bao Y, Jiang Y, et al. Diagnosis of hereditary spherocytosis and secondary hemochromatosis in a patient with jaundice. Acta Haematol. 2018;139:168–70.
Fujino T, Inoue S, Katsuki S, Higo T, Ide T, Oda Y, et al. Fatal cardiac hemochromatosis in a patient with hereditary spherocytosis. Int Heart J. 2018;59:427–30.
Hoblinger A, Erdmann C, Strassburg CP, Sauerbruch T, Lammert F. Coinheritance of hereditary spherocytosis and reversibility of cirrhosis in a young female patient with hereditary hemochromatosis. Eur J Med Res. 2009;14:182–4.
Zimelman AP, Miller A. Primary hemochromatosis with hereditary spherocytosis. Arch Intern Med. 1980;140:983–4.
Barry M, Scheuer PJ, Sherlock S, Ross CF, Williams R. Hereditary spherocytosis with secondary haemochromatosis. Lancet. 1968;2:481–5.
Ichiche M, Lacor P, Hoorens A, Vanden Brande J, Brussaard H, Vanstraelen D. Congenital spherocytosis with hereditary hemochromatosis without pathogenic mutations in the HFE gene. Eur J Intern Med. 2004;15:460–2.
Kizhatil K, Yoon W, Mohler PJ, Davis LH, Hoffman JA, Bennett V. Ankyrin-G and beta2-spectrin collaborate in biogenesis of lateral membrane of human bronchial epithelial cells. J Biol Chem. 2007;282:2029–37.
Ipsaro JJ, Mondragon A. Structural basis for spectrin recognition by ankyrin. Blood. 2010;115:4093–101.
Ipsaro JJ, Huang L, Mondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood. 2009;113:5385–93.
Lux SE, John KM, Kopito RR, Lodish HF. Cloning and characterization of band 3, the human erythrocyte anion-exchange protein (AE1). Proc Natl Acad Sci USA. 1989;86:9089–93.
Sanchez-Lopez JY, Camacho-Torres AL, Ibarra B, Tintos JA, Perea FJ. Analysis of the SLC4A1 gene in three Mexican patients with hereditary spherocytosis: report of a novel mutation. Genet Mol Biol. 2010;33:9–11.
Reithmeier RA. A membrane metabolon linking carbonic anhydrase with chloride/bicarbonate anion exchangers. Blood Cells Mol Dis. 2001;27:85–9.
Sterling D, Reithmeier RA, Casey JR. A transport metabolon. Functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J Biol Chem. 2001;276:47886–94.
Reithmeier RA, Casey JR, Kalli AC, Sansom MS, Alguel Y, Iwata S. Band 3, the human red cell chloride/bicarbonate anion exchanger (AE1, SLC4A1), in a structural context. Biochim Biophys Acta. 2016;1858:1507–32.
Acknowledgements
We appreciate the family members who participated in this study. This work was supported by the National Natural Science Foundation of China (grant number 81803562 and 81700115), Major Special Projects of Tianjin Science and Technology Service (grant number 17ZXFWGX00110), and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant number 2017-I2M-3-018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qin, L., Nie, Y., Zhang, H. et al. Identification of new mutations in patients with hereditary spherocytosis by next-generation sequencing. J Hum Genet 65, 427–434 (2020). https://doi.org/10.1038/s10038-020-0724-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0724-z
This article is cited by
-
Changes in expression levels of erythrocyte and immune-related genes are associated with high altitude polycythemia
BMC Medical Genomics (2023)
-
Genetic mutation analysis of hereditary spherocytosis in Guangxi Zhuang Autonomous Region
Journal of Hematopathology (2023)
-
Atypical Course of Hereditary Spherocytic Anemia With Severely Elevated Liver Enzymes
Indian Pediatrics (2023)
-
A novel variant of SLC4A1 for hereditary spherocytosis in a Chinese family: a case report and systematic review
BMC Medical Genomics (2022)
-
Experimental Babesia rossi infection induces hemolytic, metabolic, and viral response pathways in the canine host
BMC Genomics (2021)