Abstract
A decade ago, we described novel de novo submicroscopic deletions of chromosome 14q11.2 in three children with developmental delay, cognitive impairment, and similar dysmorphic features, including widely-spaced eyes, short nose with flat nasal bridge, long philtrum, prominent Cupid’s bow of the upper lip, full lower lip, and auricular anomalies. We suggested that this constituted a new multiple congenital anomaly—intellectual disability syndrome due to defects in CHD8 and/or SUPT16H. The three patients in our original cohort were between 2 years and 3 years of age at the time. Here we present a fourth patient and clinical updates on our previous patients. To document the longitudinal course more fully, we integrate published reports of other patients and describe genotype–phenotype correlations among them. Children with the disorder present with developmental delay, intellectual disability, and/or autism spectrum disorder in addition to characteristic facies. Gastrointestinal and sleep problems are notable. The identification of multiple patients with the same genetic defect and characteristic clinical phenotype, confirms our suggestion that this is a syndromic disorder caused by haploinsufficiency or heterozygous loss of function of CHD8.
Similar content being viewed by others
Introduction
Ten years ago, we established that submicroscopic deletions of 14q11.2 cause a characteristic clinical presentation including developmental delay, cognitive impairment, and a recognizable facies, which we suggested was syndromic [1]. Subsequently others [2, 3] have reported case studies of patients with the same genetic defect and the corresponding phenotype. Similarly, defects in CHD8, one of our two suggested candidate genes [1] have also been reported in patients who shared at least some of the same clinical features as our index patients, giving rise to the notion that the same syndrome may be caused by either CHD8 haploinsufficiency or heterozygous loss-of-function sequence mutations [4,5,6,7,8]. We review these publications, collating all patients reported to date and conducting an extensive comparison among them. We add a clinical update on our two initial Vancouver index patients and present a detailed longitudinal history for a new additional patient who was diagnosed in adulthood. Taken together, the overlap of clinical features among reported patients strongly support the conclusion that this is indeed a novel syndrome, and further we show that its’ neurocognitive profile spans developmental delay, intellectual disability (ID), autism spectrum disorder (ASD), and related neurobehavioural phenotypes [1].
Patients and methods
Patients and families were enrolled in the study of rare diseases project at the BC Children’s and Women’s Hospital. This study is approved by the BC Children’s and Women’s hospital research ethics boards (Institutional Review Board # H09-01228). All families (parents for themselves and on behalf of children) provided informed consent to participate.
Patient ascertainment methods
The novel patient reported here, Vancouver 4444, was first referred for standard clinical microarray by consultant clinician. Whole-genome microarray analyses using Affymetrix CytoChip 2.6 arrays did not yield any copy number variants (CNVs) above clinical reporting threshold. However, owing to the personal interest of the clinician a manual curation was conducted and showed a 33-Kb microdeletion spanning 54 probes (Figure S1) that overlapped the critical region reported in our first paper [1]. We then validated this CNV using quantitative polymerase chain reaction (qPCR) as previously described [9]. Primers used are given (Table S2).
Gene expression analyses
Gene expression analyses for candidate genes was conducted using quantitative gene expression assay as follows: total RNA from PBMCs was extracted using TRIzol (ThemoFisher, MA, USA) and the RNeasy Micro Kit (QIAgen, Germany). RNA was converted to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, CA, USA). qPCR reaction was set-up using GoTaq qPCR Master Mix (Promega, WI, USA) and primers (Table S2) on ViiA 7 Real-Time PCR System (ThermoFisher, MA, USA). Fold changes of gene expression were calculated using ∆∆CT method and normalized based on 18s rRNA values.
Patient identification and collation methods
Patients were identified using PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and Google Scholar (https://scholar.google.com/) searches for published reports of patients with lesions overlapping our first reported critical region, as well as involving either one or both candidate genes [1], and searching in DECIPHER (https://decipher.sanger.ac.uk/), ClinGen (https://www.clinicalgenome.org/, which includes searches in ISCA, NCBI), and the UCSC genome browser (https://www.genome.ucsc.edu) for both CNVs (≤5 Mb in size) and single-nucleotide variants (SNVs) that affect both candidate genes suggested in our original paper [1]. Where the same patient has been reported in multiple sources, clinical details were collated from all sources, with publications taking precedence over database records. Please see supplementary table (Table S1) for details.
Results
Vancouver patient 4444—new patient
This patient was the product of an uncomplicated pregnancy. Delivery was by Caesarean section due to breech presentation. There were no perinatal or prenatal complications. Birth weight was 4.8 kg. A brother, sister, and paternal half-sister are reportedly well. Family history is negative for diagnosed ASD or ID. His mother suffers from anxiety and depression. He was adopted at an early age, but both parents maintain contact. He sat at 6–7 months, walked at 19 months, and was not toilet trained until 4 years of age. At age 2 and a half years, he was diagnosed with ASD and developmental delay. He went on to manifest mild receptive and severe expressive language disability.
He suffered early separation anxiety, rocked from an early age, and later developed generalized anxiety disorder, as well as what was described as obsessive–compulsive disorder, with compulsions that include counting stripes on clothing, significant skin picking, trichotillomania, wiping after defecation to the point of inducing bleeding, and ear cleaning with Q-tips to the point of injury. Fluvoxamine was mildly helpful but caused akathisia. He has a long-standing history of fixating and perseverating on phrases and actions of others, which he will repeat frequently for weeks or months at a time. For about a year, he induced vomiting regularly. He also manifests lifelong issues with episodic insomnia, agitation, and aggression towards others, particularly if attempts are made to interrupt his rituals. These symptoms were worsened by Sertraline, Quetiapine, and Lorazepam but improved by Escitalopram, Clonazepam, and Risperidone.
Notably, after being simultaneously started on two ‘nutraceutical’ supplements (trehalose and Sagee) [10] without medical advice, he had an immediate and paradoxically severe adverse reaction. He developed severe insomnia, impaired concentration, and dystonic deviation of his face and jaw that was refractory to benztropine treatment. He returned to his baseline profile after discontinuation of these supplements.
A generalized tonic-clonic seizure occurred at age 21 years soon after Escitalopram was added to Risperidone, moments after he had been straining to pass stool for a prolonged period, raising the possibility of convulsive syncope. Several waking and sleep electroencephalograms, computed tomographic and magnetic resonance imaging scans, and Holter monitor were unremarkable.
Patient 4444 was assessed clinically and neuropsychiatrically at 22 years of age. On examination, height was 175 cm (16th percentile). He was non-obese. Head circumference was 60 cm (99th percentile for height). Inner canthal, interpupillary, and outer canthal distances were 3.5 cm (90th percentile), 6 cm (67th percentile), and 9 cm (60th percentile), respectively. Palpebral fissures were downslanting, with a width of 2.5 cm (1st percentile) bilaterally. The face had a triangular appearance. His forehead and supraorbital ridges were prominent. There was prognathism and a broadened chin, and a large nose, with high nasal bridge, upslanting base, anteverted nares, and broad triangular tip. Pinnae were low-set and posteriorly rotated. Mild left facial and chest hypoplasia, webbing of the neck, and rhizomelic shortening of his arms (34 cm) were noted. Hands were broad with short palms, fingers, and thumbs, with short distal phalanges and broad nail bases. There was mild syndactyly involving all interspaces in the hands (Fig. 1).
Composite of patient photographs. Composite figure showing photographs of the three patients reported here, Vancouver #5566, #8356, and #4444, as well as images from publications by Bernier et al. [6], Drabova et al. [3], Merner et al. [4], Prontera et al. [2], and Stolerman et al. [5]. Permissions have been obtained from the publishers for reproduction of images previously published
Although poorly engaged by the examination, he did not appear distracted by internal or external stimuli. He recited the days of the week forwards with difficulty but could not do so backwards. Speech was very terse and limited and delivered in an unusual, high-pitched, hypernasal voice, with frequent idiosyncratic upward tonal inflection at the end of phrases. There was obvious difficulty establishing and switching cognitive set. Despite prominent echolalia and echopraxia, other catatonic signs were absent. Neurological examination was notable for prolonged jaw opening (which may be due to emulation of another day programme client), occasional lip pouting, and some lower facial and very minor lingual dyskinesias. There was no retrocollis.
Clinical chromosomal microarray analysis using Affymetrix Cytoscan HD did not yield any variants meeting clinical reporting thresholds. However, manual curation revealed a 33.269 kb deletion (called by 56 contiguous probes) extending from co-ordinates 21,827,942 to 21,861,211 (hg19) in the minimally involved region (Fig. 2), which is almost exactly the critical region we first reported in 2007 [1]. The CNV was verified as de novo heterozygous by qPCR. qPCR CHD8 expression analysis confirmed reduced gene expression versus his normal brother (age- and sex-matched control).
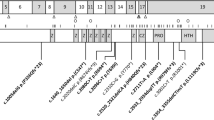
UCSC Genome browser view of the affected region (GRCh37/Hg19). The top box contains a view of the region affected by the largest copy number variant (CNV) reported involving the critical region, while the bottom box contains a zoomed-in view of CHD8 only, showing placement of all damaging single-nucleotide variants (SNVs) reported within the gene. Top box: top panel contain the following standard and custom tracks; RefSeq genes, CNV/SNVs in the region (custom track), and CNVs in normal individuals reported in the Database of Genomic Variation. The region containing CHD8 has been highlighted by a light blue vertical bar. Bottom box: CHD8 and all SNVs falling within the gene, reported by Bernier et al. [6]
Vancouver patient 5566—update
Vancouver patient 5566 whom we originally described at age 28 months [1] was reassessed at 12 years of age to provide a longitudinal update. She has been diagnosed with mild ID, attending grade 6 with an individualized education plan. She writes at a grade 1 or 2 level, and her printing is at a kindergarten level. She does have some difficulties with fine motor and gross motor skills. She was assessed for ASD but did not meet full diagnostic criteria. She has been diagnosed with an anxiety disorder, with little improvement after 6 years of psychological therapy. Particular triggers include burned-out light bulbs and bathtub water jets. On examination, her height was 156.8 cm (75th percentile), weight 49.7 kg, body mass index (BMI) 20.2 kg/m2 (77th percentile), and head circumference 56.6 cm (98th percentile). Head and neck examination revealed prominent and deep-set eyes, with an inner canthal distance of 2.7 cm (25th %ile), interpupillary distance of 5.6 cm (75th %ile), and palpebral fissures measuring 2.7 cm bilaterally (~10th %ile). She had hypernasal speech, and a prominent cupid’s bow of the mouth. She had a high-arched palate with some dental malocclusion. Her enamel appeared mottled and very thin. She also had a crease between her lower lip and the end of her chin, similar to that of patients with Weaver syndrome (Fig. 1). On examination of the thorax, she exhibited a right rib prominence when bending over. Subsequent independent radiograph assessments showed scoliosis that progressed from 23 degrees at 12 years of age up to 33 degrees at 14 years of age. Breast development was Tanner stage III, while pubic hair was Tanner stage III in an adult distribution. She has documented sleep problems since 4 years of age. She continued to exhibit truncal hypotonia, with a knock-kneed appearance on standing. When standing unsupported, she did not fully straighten her knees. There was no true muscle weakness, but decreased tone was noted in both arms and legs. Video 1 provides a film of her gait.
Vancouver patient 8326—update
Patient 8326, originally reported at 44 months of age [1], was re-assessed at age 11 years and 8 months. He initially presented with global developmental delay, with severely affected speech. He continued to have limited verbal communication. At age 4 years, he was diagnosed with ASD (at the time, falling under ‘pervasive developmental disorder—not otherwise specified), and mild ID. At the time of reassessment, he had been reading and writing at a grade 3–4 level and performing mathematics at grade 3 level. He was also diagnosed with an anxiety disorder, presenting with selective mutism. He has a history of atypical febrile seizures, with a total of 5–6 seizures between the ages of 2 and 10 years. He has not required antiepileptic medications. Previous electroencephalograms demonstrated rare bursts of sharp waves in the right posterior quadrant during sleep. With regards to dentition, he had poor dental enamel of both primary and permanent teeth, a number of extra permanent teeth and dental crowding, and multiple caries, with dental treatment first required at age 2 years. With respect to gastrointestinal (GI) issues, chronic constipation has been noted.
On examination, his height was 148.5 cm (50th percentile), weight 32 kg, BMI 14.5 kg/m2, and head circumference 53 cm (25th–50th percentile). His inner canthal distance measured 3.8 cm (95th percentile), and inter pupillary distance was 5.8 cm (75th percentile). His philtrum measured 2 cm, and he demonstrated a prominent cupid’s bow. He had a narrow high-arched palate, with significant teeth crowding, more pronounced in the lower teeth (Fig. 1). Each pinna had a prominent anti-helix, prominent crus of the helix, and inferior crus of the inferior antihelix. He also exhibited a small ear pit anterior to the ascending part of the left helix, similar to that of his sister and father. He has mild bilateral 5th finger clinodactyly, planus valgus, and mild varus deformity of the first toe bilaterally. Video 1 provides a film of his gait.
Discussion
Existence of a distinct syndrome is confirmed
In the decade since first reporting the condition, several studies have reported patients who have lesions involving the candidate genes we suggested. We have collated all clinical information on published and reported patients as Table S1, totalling 51 patients. There are 27 published patients (Table S1) [2,3,4,5,6,7,8, 11], including the 4 patients we report (3 in our original paper and 1 in this report). Nine of the 27 published patients appear as case studies, while 17 patients were identified by large cohort studies in ASD, due to presence of SNVs and Indels in CHD8. Of these, 15 patients’ variation is confirmed to be pathogenic, and we include them in our detailed phenotypic comparison along with the 9 case reports (Fig. 3). As of November 2017, six patients are reported in the ClinGen database and seven in the DECIPHER database to have CNVs involving our critical region (we only consider lesions <5 MB as a CNV). Eleven reported patients in DECIPHER have SNVs in CHD8, and we collate them in Table S1. In contrast, there are far fewer reports implicating SUPT16H [MIM: 605012]; the gene is located within CNVs in patients published as case reports or collected in ClinGen and DECIPHER as described below; in all cases however, the CNV includes both SUPT16H and CHD8 [2, 3]. However, to our knowledge, no studies have been published reporting causative variation in only SUPT16H; there are only two SNVs reported in DECIPHER within this gene, of which only one (DECIPHER 323372) is annotated as likely pathogenic and which we have included in our detailed patient comparison table (Table S1). Finally, since our suggested critical region [1] is included in clinical targeted array platforms, we expect more patients to be identified. Indeed, we know several families who have posted to a closed social media support group for this condition. We have excluded two case reports as the large size of their CNVs precludes a meaningful comparison: Smyk et al. [12] described a microduplication encompassing both genes in a larger region. Terrone et al. [13] report a patient with features of Wolf–Hirschorn syndrome (WHS), with a much larger deletion that possibly involves the WHS region.
Schematic representation of common features. Illustrates the most common phenotypes between reported cases for this syndrome. Each circle is a separate phenotype. Circles are as follows; G.I- gastrointestinal complications, Motor- motor complications, Skeletal- skeletal complications, P.P- pes planus. The other circles are placed in three main groups. Group one, collects the most common phenotypes between cases (Macro- macrocephaly, ASD- autism spectrum disorder, DD- developmental delay, ID- intellectual disability). Group two collects neurological-related complications (Neuro- neurological complications, Speech- speech delay, O.B- other behavioural complications, Sleep- sleep problems). Group three collects craniofacial dysmorphisms (Face- facial dysmorphia, Ears- auricular anomalies, H.P- high palate, Dental- dental problems). Each segment within a circle represents a patient, the 9 patients from the case reports are organized as the top half of the circle (‘Z’ Zheng et al., 2015 [11]; ‘126’ Zahir et al., 2007 [1]; ‘S’ Stolerman et al., 2016 [5]; ‘P’ Prontera et al., 2014 [2]; ‘977’ current study and Zahir et al., 2007 [1]; ‘976’ current study and Zahir et al., 2007 [1]; ‘D’ Drabova et al., 2014 [3]; ‘M’ Merner et al., 2015 [4]; and ‘4444’ current study), while the 15 patients from the ASD cohort study by Bernier et al. [6] are organized as the bottom half of the circle. The colouring of each segment is as follows; green: presence of the phenotype when the phenotype is reported as ‘+’ only, blue: presence of the phenotype when the phenotype is described (Table S1), white: absence of phenotype (tested and found absent), while grey: phenotype is not reported (we are unsure if it present or absent). The length of each segment represents the age of the case when last reported (3–41 years) and is in ascending order
In order to better determine the clinical spectrum for this disorder, we have conducted a careful comparison between the 24 patients for whom we have detailed clinical information. A careful comparison of the facial gestalt and other clinical features for patients with the ASD ‘subtype’ [6] and those reported by us and others [1,2,3,4,5, 11] indicate the disorder is one and the same (Figs. 1 and 3). Developmental delay, ID, ASD, other behavioural defects, macrocephaly, the facial gestalt, speech defects, GI problems, and motor defects appear the most distinguishing features.
Nevertheless, non-recurrent phenotypes are reported; Patient Vancouver 5566 exhibited significant scoliosis that progressed by 10 degrees over 2 years in early adolescence. Dental issues have been reported for Vancouver patients 5566 and 8326, and for patients reported by Prontera et al. and Drabova et al. (Fig. 1). Structural deficits for the ears, hands, and feet for the patients reported via case studies are not noted in the ASD reports, probably because assessments for ASD do not typically detail non-behavioural phenotypes. Similarly, while almost all patients from the ASD reports are noted to have sleep problems, this is not the case for patients from the case studies where questions about sleep patterns might not have been asked.
Finally, we note a patient reported by Kimura et al. [14] who was found to have a SNV in CHD8 that the authors designate as causative (Table S1). What is remarkable about this patient is that the major diagnosis was of schizophrenia at age 20 years. The patient also reported persistent GI problems, which may have led to death at age 67 years. Repetitive and catatonic behaviours noted in this patient are similar to those observed in our patient Vancouver 4444, who has presented schizoid-like behaviours. However, as information verifying the pathogenicity of the SNV is sparse and as the patient was identified by a targeted screen for mutations in only four known causative genes, we are unsure whether other undiscovered causative factors may play a role in this case, nevertheless we note that this report possibly expands the CHD8 causative profile into schizophrenia.
CHD8 is a syndromic ID/ASD causative locus
In our patients, the distal end of CHD8 and a region greater than the proximal half of SUPT16H are involved (Fig. 2), whose candidacy we discuss below.
CHD8
CHD8 (OMIM# 610528) is an SNF2-like member of the chromodomain helicase DNA-binding protein family, which has widespread and complex effects on gene transcription. CHD8 is ubiquitously expressed with highest expression levels in the cerebellum and is expressed in both fetal and adult humans. Current evidence has mounted in support of CHD8 as a ‘master driver’ in ASD [15] with SNVs in CHD8 shown to be a significant cause of ASD, explaining up to 0.4% of cases in a cohort of >3000 children [6]. Point mutations in CHD8 are reported to cause a ‘subtype’ of ASD characterized by macrocephaly and GI problems [6, 15]. Chd8+/− mice have increased brain volume, anxiety, and autistic-like behaviours, as well as GI defects, while homozygous knockout mice are embryonic lethal [16]. An increase in intra-ocular distance, as well as increase in total brain volume, and behavioural deficits were noted in Chd8+/− mice versus wild-type littermates [17]. Zebra fish studies showed increased head size and gut defects for chd8-suppressed morpholinos [6]. These studies support the notion that haploinsufficiency of the gene is sufficient to cause at least the ASD, macrocephaly, and GI phenotypes.
Also noteworthy is the fact that CHD8 has been recently recognized as a prognostic indicator for gastric cancer [18] among other cancers of endodermal tissues [19, 20], and patient Nij07-06646 from the Bernier et al. study [6] who carries a paternally inherited splice site mutation in CHD8 has a family history remarkable for death of father due to complications from a renal carcinoma that was first detected at 42 years of age (Table S1). The father showed similar autistic features as patient Nij07-06646. Another remarkable finding is that in a recent study of individuals with overgrowth and ID, CHD8 was among the six genes that accounted for 44% of variations found in 710 patients [21], which is relevant because macrocephaly, an overgrowth phenotype, is a distinguishing feature of this syndrome (Fig. 3), while among our cohort we note that 9 of the 15 children with reported heights have a height ≥+ 2 SD (Table S1).
SUPT16H
In contrast, there is far less information on SUPT16H in order to robustly determine whether it plays any role in the phenotype. SUPT16H encodes the larger subunit of the FACT (Facilitates Chromatin Transcription) complex, a chromatin remodeler with histone chaperone function that plays a role in diverse cellular processes [22, 23]. FACT has also been shown to play a key role in DNA transcription by modulating DNA–chromatin interaction [24], including that involving histone variants [25]. Other than the patients with CNVs that involved both CHD8 and SUPT16H, only one other case is reported to our knowledge, who carries a damaging variant in SUPT16H: patient 323372 recorded in DECIPHER. This patient carries a SNV in SUPT16H that may be pathogenic, with six recorded phenotypes: ASD, severe ID, precocious puberty, proportionate growth increase, wide nasal bridge, and cupid’s bow of the upper lip (Table S1).
CHD8 and SUPT16H expression in patient 4444
Gene expression analysis in patient Vancouver 4444 showed that the expression was reduced for both CHD8 (0.5745 ± 0.04556, n = 3, P value = 0.0050) and SUPT16H (0.6122 ± 0.03159, n = 3, P value = 0.0359) versus his normal brother, who served as an age- and sex-matched control (Fig. 4). A protein interaction analysis for CHD8 and SUPT16H shows that they have a high co-expression profile (score 0.089, Stringdb.org, version 10.5). Preliminary evidence suggests a possible role for both CHD8 and SUPT16H to influence gene expression via RNApol II elongation activity [26]. However, any involvement of SUPT16H may only be anecdotal, due to the gene’s position next to CHD8. Alternatively, as the products of both genes are active in epigenetic pathways, a modifier role for SUPT16H can be hypothesized.
Remarkably, of the 51 patients we collect in Table S1, only 7 carry variation that includes SUPT16H, of which six have CNVs that include both CHD8 and SUPT16H. Only one patient, patient #323372 reported in DECIPHER, has a lesion in only SUPT16H; there is no published information on this patient, and annotating the variant ourselves, we find conflicting evidence with respect to its pathogenicity. Further, there are now a number of molecular functional studies that establish the causativeness of CHD8 [6, 16, 17, 27, 28]. The evidence thus points to haploinsufficiency of CHD8 as being highly penetrant for this syndrome, nevertheless it is premature to rule out a contribution from SUPT16H.
CHD8 expression directs possible drug repurposing
Patient Vancouver 4444’s response to psychotropic drugs and nutraceuticals may have pharmacotherapeutic implications. Notably, perseveration, rigidity, and expressive language function deteriorated significantly on Divalproex (a valproic acid congener). Valproic acid significantly downregulates the insulator CTCF [29], whereas knockdown of CHD8 results in reduced function at CTCF-binding sites[30], suggesting that the drug could worsen any adverse epigenetic CHD8 haploinsufficiency impact. The nutraceuticals trehalose and sagee caused severe insomnia, impaired concentration, and facial and jaw dystonia. Trehalose is thought capable of inducing autophagic degradation [31]. Autophagy is limited by β-catenin, and DMBT, a trehalose derivative, may inactivate β-catenin signalling [32]. CHD8 negatively regulates β-catenin via direct binding and also downregulates β-catenin-responsive genes [33]. Haploinsufficiency of CHD8 may have therefore potentiated the autophagy-promoting effects of trehalose. The 5-alpha reductase inhibitor finasteride, used to treat benign prostatic hypertrophy and alopecia androgenita, upregulates CHD8 expression [34] and attracts potential treatment interest. Intriguingly, the amphetamine MDMA (‘Ecstasy’), which is being studied in a registered clinical trial for ASD [35], also upregulates CHD8 expression [34].
A distinct genetic disorder with ID and/or ASD
A decade ago, we suggested overlapping microdeletions in 14q11.2 that included CHD8 and SUPT16H in the critical region may be syndromic [1]. Here we establish this new syndrome concluding that haploinsufficiency or loss-of-function mutations of CHD8 produce a distinct neurodevelopmental syndrome, with a cognitive and behavioural profile beginning with developmental delay, progressing to ID and/or ASD, and with other neurofunctional features, such as anxiety. Characteristic facial features include hypertelorism, a long philtrum, down-slanting palpebral fissures, and macrocephaly. Feeding issues are often noted in infancy and GI dysfunctions are common, as are sleep disturbances.
The mutation spectrum of CHD8 mirrors that of its family member CHD7, where CNVs involving CHD7 were first identified in CHARGE syndrome patients and subsequently SNVs in the gene established it as causative. Similarly, we first reported CNVs involving CHD8 and subsequently CHD8 SNVs have been found to cause the same disorder, which in this case is a newly identified syndrome. With respect to SUPT16H, we cannot rule out a contributory role that may expand the phenotypic spectrum. Given the association of CHD8 mutations with cancer, we also recommend that early cancer screening be conducted for patients with this new recognizable syndrome spanning neurodevelopmental disorders.
References
Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44:556–61.
Prontera P, Ottaviani V, Toccaceli D, Rogaia D, Ardisia C, Romani R, et al. Recurrent approximately 100 Kb microdeletion in the chromosomal region 14q11.2, involving CHD8 gene, is associated with autism and macrocephaly. Am J Med Genet A. 2014;164A:3137–41.
Drabova J, Seemanova E, Hancarova M, Pourova R, Horacek M, Jancuskova T, et al. Long term follow-up in a patient with a de novo microdeletion of 14q11.2 involving CHD8. Am J Med Genet A. 2015;167:837–41.
Merner N, Forgeot d’Arc B, Bell SC, Maussion G, Peng H, Gauthier J, et al. A de novo frameshift mutation in chromodomain helicase DNA-binding domain 8 (CHD8): a case report and literature review. Am J Med Genet A. 2016;170A:1225–35.
Stolerman ES, Smith B, Chaubey A, Jones JR. CHD8 intragenic deletion associated with autism spectrum disorder. Eur J Med Genet. 2016;59:189–94.
Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell . 2014;158:263–76.
O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50.
Tucker T, Zahir FR, Griffith M, Delaney A, Chai D, Tsang E, et al. Single exon-resolution targeted chromosomal microarray analysis of known and candidate intellectual disability genes. Eur J Hum Genet. 2014;22:792–800.
Sagee (Natural Product Number 80031803) [Internet]. Health Canada. 2016 [cited 23 Apr 2017]. Available from: https://health-products.canada.ca/lnhpd-bdpsnh/search-recherche.do;jsessionid=0AF7AC4540CF9F9F9015CADDC894644D.
Zheng Q, Zhan G, Tang C, Bi X, Zhao W, Zou X. A novel CHD8 gene mutation in a patient with autism spectrum disorder and literature review. Chin J Evid Based Pediatr. 2015;10:216–21.
Smyk M, Poluha A, Jaszczuk I, Bartnik M, Bernaciak J, Nowakowska B. Novel 14q11.2 microduplication including the CHD8 and SUPT16H genes associated with developmental delay. Am J Med Genet A. 2016;170A:1325–9.
Terrone G, Cappuccio G, Genesio R, Esposito A, Fiorentino V, Riccitelli M, et al. A case of 14q11.2 microdeletion with autistic features, severe obesity and facial dysmorphisms suggestive of Wolf-Hirschhorn syndrome. Am J Med Genet A. 2014;164A:190–3.
Kimura H, Wang C, Ishizuka K, Xing J, Takasaki Y, Kushima I, et al. Identification of a rare variant in CHD8 that contributes to schizophrenia and autism spectrum disorder susceptibility. Schizophr Res. 2016;178:104–6.
Barnard RA, Pomaville MB, O’Roak BJ. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology. Front Neurosci. 2015;9:477.
Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–9.
Platt RJ, Zhou Y, Slaymaker IM, Shetty AS, Weisbach NR, Kim JA, et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017;19:335–50.
Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, et al. CHD8 is an independent prognostic indicator that regulates Wnt/beta-catenin signaling and the cell cycle in gastric cancer. Oncol Rep. 2013;30:1137–42.
Damaschke NA, Yang B, Blute ML Jr., Lin CP, Huang W, Jarrard DF. Frequent disruption of chromodomain helicase DNA-binding protein 8 (CHD8) and functionally associated chromatin regulators in prostate cancer. Neoplasia . 2014;16:1018–27.
Jones DH, Lin DI. Amplification of the NSD3-BRD4-CHD8 pathway in pelvic high-grade serous carcinomas of tubo-ovarian and endometrial origin. Mol Clin Oncol. 2017;7:301–7.
Tatton-Brown K, Loveday C, Yost S, Clarke M, Ramsay E, Zachariou A, et al. Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am J Hum Genet. 2017;100:725–36.
Maluchenko NV, Chang HW, Kozinova MT, Valieva ME, Gerasimova NS, Kitashov AV, et al. [Inhibiting the pro-tumor and transcription factor FACT: mechanisms]. Mol Biol (Mosk). 2016;50:599–610.
Belotserkovskaya R, Reinberg D. Facts about FACT and transcript elongation through chromatin. Curr Opin Genet Dev. 2004;14:139–46.
Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–79.
Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, Robert F. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol Cell. 2015;58:1113–23.
Rodriguez-Paredes M, Ceballos-Chavez M, Esteller M, Garcia-Dominguez M, Reyes JC. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009;37:2449–60.
Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–77.
Wang P, Lin M, Pedrosa E, Hrabovsky A, Zhang Z, Guo W, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism. 2015;6:55.
Jergil M, Forsberg M, Salter H, Stockling K, Gustafson AL, Dencker L, et al. Short-time gene expression response to valproic acid and valproic acid analogs in mouse embryonic stem cells. Toxicol Sci. 2011;121:328–42.
Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42.
Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52.
Tang L, Yue B, Cheng Y, Yao H, Ma X, Tian Q, et al. Inhibition of invasion and metastasis by DMBT, a novel trehalose derivative, through Akt/GSK-3beta/beta-catenin pathway in B16BL6 cells. Chem Biol Interact. 2014;222:7–17.
Nishiyama M, Skoultchi AI, Nakayama KI. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-beta-catenin signaling pathway. Mol Cell Biol. 2012;32:501–12.
Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45(D1):D972–8.
Danforth AL, Struble CM, Yazar-Klosinski B, Grob CS. MDMA-assisted therapy: a new treatment model for social anxiety in autistic adults. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:237–49.
Acknowledgements
Our special thanks to the families who readily provided updated photographs and video and who have been involved participants all along.
Funding
FRZ was supported by a CIHR Post-Doctoral Scholarship, NeuroDevNet post-fellowship, and UBC BlumaTischler fellowship. WTG is supported by a BCCHR Intramural IGAP award and by Canadian Institutes of Health Research (CIHR) Grant PJT-148830. This work was supported by funds from the CIHR grant MOP-102600.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yasin, H., Gibson, W.T., Langlois, S. et al. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J Hum Genet 64, 271–280 (2019). https://doi.org/10.1038/s10038-019-0561-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0561-0
This article is cited by
-
CHD8 mutations increase gliogenesis to enlarge brain size in the nonhuman primate
Cell Discovery (2023)
-
Clinical evaluation of rare copy number variations identified by chromosomal microarray in a Hungarian neurodevelopmental disorder patient cohort
Molecular Cytogenetics (2022)
-
Yield of clinically reportable genetic variants in unselected cerebral palsy by whole genome sequencing
npj Genomic Medicine (2021)
-
Next-generation gene panel testing in adolescents and adults in a medical neuropsychiatric genetics clinic
neurogenetics (2021)
-
Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology
Nature Communications (2021)