Abstract
A wet-chemical approach using DNA extracted from salmon milt as a template to mass produce Ag nanoparticles was developed. Spherical Ag nanoparticles with a main diameter of less than 10 nm were obtained. The concentration of Ag nanoparticles in the as-produced colloidal suspension was as high as 5.3 × 10−2 mol l−1. This simple and effective procedure should offer an alternative route to the mass production of Ag nanoparticles for practical applications.
Similar content being viewed by others
Introduction
Many potential applications for the remarkable electrical and chemical properties of silver (Ag) nanoparticles have been proposed.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 The so-called wet-chemical reduction method has been the standard technique for the production of Ag nanoparticles because of its simplicity and high production efficiency. In most wet-chemical reduction methods, biopolymers such as polysaccharides,11, 12, 13 proteins14 and nucleic acids7, 15, 16, 17, 18, 19 have been used. These biopolymers function as templates for the desired nano-sized structures of Ag nanoparticles. DNA, the most important biopolymer, is also capable of forming Ag nanoparticles through the formation of DNA–Ag(I) complexes. DNAs with a uniform molecular weight and a specific sequence (artificial oligonucleotides,7, 15, 16, 17 plasmid DNA18 and bacteriophage T4 DNA19) have been commonly used. However, there are difficulties in the mass production of these types of DNA.
In our previous studies, we demonstrated that DNA could be obtained on an industrial scale by using salmon milt as the DNA source.20, 21, 22, 23 In this study, we have evaluated the possibility of using the salmon milt-based DNA as a template for the mass production of Ag nanoparticles. Three types of salmon milt-based DNA were used, each differing in molecular weight. DNA with an average molecular weight of ∼20 000 Da (single stranded) proved to be optimal for the reproducible production of Ag nanoparticles.
Expermental procedure
Materials
Three types of salmon milt-based DNA were used throughout this study: (i) low-molecular weight DNA (LMw-DNA; molecular weight ∼20 000 Da, single-stranded), (ii) intermediate-molecular weight DNA (IMw-DNA; molecular weight ∼50 000–100 000 Da, mostly single-stranded) and (iii) high-molecular weight DNA (HMw-DNA; molecular weight over ∼10 000 000 Da, double-stranded). These DNAs were obtained from Nissei Bio Co., Ltd., Eniwa, Japan. The molecular weights of the DNAs were estimated by HPLC gel filtration for the LMw-DNA and gel electrophoresis for the IMw-DNA and HMw-DNA. The determination of double-stranded or single-stranded DNA was made by measuring the increase in absorbance at 260 nm upon heating the DNA solutions. Silver nitrate (AgNO3, special grade) was purchased from Kishida Chemical Co., Ltd. (Osaka, Japan). Aqueous ammonia (10%), sodium borohydride (NaBH4, chemical grade) and nitric acid (HNO3, for analysis of poisonous metal grade) were obtained from Wako Pure Chemicals Industries Ltd. (Osaka, Japan). Potassium peroxydisulfate (for N and P analysis grade) was purchased from Kanto Chemical Co., Inc. (Tokyo, Japan). Ultra-pure water (supplied from Milli-Q water purification systems, Merck Millipore, Billerica, MA, USA, >18 MΩ) was used in all experiments.
Preparation of the Ag nanoparticles
The reactions of DNA (5 × 10−5 mol l−1 nucleotide) with Ag(I) at a molar ratio of [nucleotide]:[Ag(I)] of 1:1-1:30 were monitored by ultraviolet-visible (UV–vis) spectroscopy (V-560, Jasco, Tokyo, Japan). DNA solutions (0.33–33 g l−1, that is, 10−3–10−1 mol l−1 nucleotide) were prepared by adding DNA to aqueous ammonia (1.0%). For the Ag nanoparticles preparation, the DNA solution (10−3 or 10−2 mol l−1) and aqueous AgNO3 (10−2 or 10−1 mol l−1) were mixed at a volume ratio of 1:2 (molar ratio of 1:20), and magnetically stirred in an ice bath. Under continuous mixing, 10−1 or 1 mol l−1 of NaBH4 in aqueous ammonia (10%) was added to the solution; the molar ratio of NaBH4 to Ag(I) was 1:1. After standing overnight, the supernatant, which included the Ag nanoparticles, was dialyzed using a dialysis membrane (molecular weight cutoff: 14 000, Viskase Companies Inc., Darien, IL, USA) for ∼24 h.
Characterization of the Ag nanoparticles
UV–vis spectroscopy was used to monitor the reactions between Ag(I), DNA and NaBH4. The sample solutions for the UV–vis measurements were diluted 500-fold with water. The morphologies of the Ag nanoparticles were evaluated by visual observation, UV–vis spectroscopy, transmission electron microscopy (TEM, JEM-2000ES, JEOL, Akishima, Japan) and scanning transmission electron microscopy (STEM, HD-2000, Hitachi, Tokyo, Japan). The samples for the TEM/STEM observations were prepared by spotting the diluted dispersion on a lacey carbon film supported by a 400-mesh copper grid (Cu-400CN, Pacific Grid-Tech, San Francisco, CA, USA) and drying at room temperature.
The concentrations of Ag, P and Na in the dispersion were measured with inductively coupled plasma atomic emission spectrometry (ICP-AES, ICPE-9000, Shimadzu, Kyoto, Japan). The samples were hydrolyzed in the presence of aqueous potassium peroxydisulfate (4.0%) at 120 °C and then diluted to an adequate concentration with HNO3 (10−1 mol l−1). The amount of Ag and DNA in the Ag nanoparticles was calculated from the concentrations measured by ICP-AES and from the solid weight after drying by heating.
The particle size and shape were observed with TEM and atomic force microscopy (AFM, picoscan2500, SII, Chiba, Japan). AFM samples were prepared by spotting the diluted dispersion on exfoliated mica and drying at room temperature.
The zeta-potential of the Ag nanoparticles in water was measured with a particle analyzer (Delsa Nano HC, Beckman Coulter, Brea, CA, USA). After confirming the dispersion of the samples by dynamic light scattering, the zeta-potential was determined by electrophoretic light scattering and automatically calculated using the Smoluchowski equation. The sample solution was diluted 10-fold with water, and a flow cell was used for the measurement.
The thermal behavior of the Ag nanoparticles was investigated by thermogravimetry (TG, TG-DTA6200, SII) from 25 to 1000 °C at a heating rate of 5 °C min−1 under an air atmosphere. In addition, the decomposition behavior of the DNA in the Ag nanoparticles was examined by a stepwise heating, maintaining each step for 4 h at 200, 300, 400, 500 and 600 °C.
Long-term stability of the Ag nanoparticles in water
An aqueous dispersion of Ag nanoparticles was stored without dilution in a plastic tube wrapped with aluminum foil to protect from light for 6 months at room temperature. The sample was then evaluated visually and by UV–vis spectroscopy.
Results and discussion
Ag nanoparticles with salmon milt-based DNA as the template
DNA is capable of selectively binding Ag(I) with their nucleotide bases; this binding changes the wavelength of the maximum absorption of DNA.15, 24, 25 In this study, the change in the absorption at 260 nm upon addition of Ag(I) (Figure 1) reveals the formation of the DNA–Ag(I) complex. The increase in absorbance observed only for the HMw-DNA (Figures 1b and c) was assumed to be induced by the denaturation of the double-stranded DNA.24 The absorption peak of the DNA shifted with increasing concentration of Ag(I), and then remained unchanged (Figure 1c). These results suggest that the binding of Ag(I) to DNA did not increase as the [Ag(I)]/[nucleotide] ratio rose from 10-fold to 20- or 30-fold, that is, only the amount of unbound Ag(I) increased.
Evolution of the maximum absorption of DNA in the presence of different concentrations of Ag(I): (a) low-molecular weight DNA (LMw-DNA) and (b) high-molecular weight DNA (HMw-DNA). (c) Plot of the absorbance and wavelength of the maximum absorption vs the molar ratio [Ag]:[nucleotide] from 1:1 to 1:30. Absorbance vs wavelength data for IMw are not shown.
Next, to optimize the volume ratio, 10−2 mol l−1 LMw-DNA solution and 10−1 mol l−1 AgNO3 solution were mixed at volume ratios of 1:1, 1:2 and 1:3, and then 1 mol l−1 NaBH4 solution was added in a molar equivalence to Ag(I). The solution with a volume ratio of 1:2 changed from clear to dark brown in color with very small precipitates. UV–vis absorption spectra of the DNA solution, the solution of DNA and AgNO3, and the solution after the reduction are shown in Figure 2. The maximum absorption at ∼260 nm is shifted after the addition of Ag(I), and the peak at 408 nm, the typical plasmon resonance band of Ag nanoparticles, appeared after the reduction. In the case of the volume ratio of 1:3, a large amount of precipitation was observed, and a very small amount of Ag nanoparticles was produced.
Ag nanoparticles were also prepared using solutions of IMw-DNA (10−2 mol l−1) and AgNO3 (10−1 mol l−1) and HMw-DNA (10−3 mol l−1) and AgNO3 (10−2 mol l−1). The molar ratio of [nucleotide]:[Ag(I)]:[BH4] was chosen to be 1:20:20. Ag concentrations in the mixed solutions were calculated to be 6.25 × 10−3 mol l−1 when using the 10−2 mol l−1 AgNO3 solution and 6.25 × 10−2 mol l−1 when using the 10−1 mol l−1 AgNO3 solution. As denoted in Table 1, homogenous dark brown solutions were obtained under all conditions. Moreover, a slight turbidity was observed without clear phase separation in the solutions with 10−2 mol l−1 IMw-DNA (Table 1, no. 4) and 10−3 mol l−1 HMw-DNA (Table 1, no. 5). Precipitation also occurred in the 10−2 mol l−1 IMw-DNA solution (Table 1, no. 4), and the Ag concentration in the supernatant was above 46% (confirmed by ICP-AES). There was little precipitation in the 10−2 mol l−1 LMw-DNA solution (Table 1, no. 2), and the Ag concentration was only ∼3%.
The dark brown supernatants were purified by membrane dialysis, a simple method amenable to mass production. After purification, all the obtained solutions were dark brown and homogeneous in appearance. The Ag concentrations as estimated by ICP-AES were 3.9 × 10−3, 4.0 × 10−3, 4.3 × 10−3, 5.3 × 10−2 and 3.6 × 10−2 mol l−1 for the 10−3 mol l−1 LMw-DNA, 10−3 mol l−1 IMw-DNA, 10−3 mol l−1 HMw-DNA, 10−2 mol l−1 LMw-DNA and 10−2 mol l−1 IMw-DNA solutions, respectively. The UV–vis absorption spectra of Ag colloids have been reported to be influenced by the particle size distribution; for example, the absorption peak shifts toward longer wavelengths as the particles become larger,26 and small aggregates cause a decrease in the peak along with the appearance of a long tail on the high wavelength side.27 At the same Ag concentration, the UV–vis absorption spectra showed a similar shape for 10−3 and 10−2 mol l−1 LMw-DNA solutions (Figure 3a, no. 1 and 2), and 10−3 mol l−1 IMw-DNA solution (Figure 3a, no. 3), suggesting that these Ag particles possessed a similar size distribution.
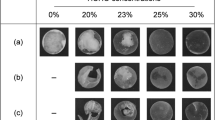
(a) Ultraviolet-visible (UV–vis) absorption spectra of the Ag nanoparticle dispersions, in terms of the same concentration of Ag. (b) transmission electron microscopy (TEM) and scanning transmission electron microscopy (STEM) images of the samples. (c) Corresponding scanning electron microscopy (SEM) images of Ag–DNA complexes after dialysis. The specified DNA solutions were: (1) low-molecular weight DNA (LMw-DNA) (10−3 mol l−1), (2) LMw-DNA (10−2 mol l−1), (3) intermediate-molecular weight DNA (IMw-DNA) (10−3 mol l−1), (4) IMw-DNA (10−2 mol l−1) and (5) high-molecular weight DNA (HMw-DNA) (10−3 mol l−1).
Figures 3b and c show typical microscopic images. The LMw-DNA at concentrations of 10−3 and 10−2 mol l−1 and the IMw-DNA at a concentration of 10−3 mol l−1 produced fine Ag nanoparticles (Figure 3a, no. 1, 2 and 3), while the IMw-DNA at a concentration of 10−2 mol l−1 and the HMw-DNA gave large Ag particles (Figure 3a, no. 4 and 5). The 10−2 mol l−1 LMw-DNA and the 10−1 mol l−1 aqueous AgNO3 solutions were selected for producing Ag nanoparticles in a high concentration.
Characterization of the Ag nanoparticles
The concentrations of Ag, P and Na in the Ag nanoparticle dispersions were measured by ICP-AES, and the DNA concentration was calculated from the amount of P. The Ag content in the nanoparticles was 85.2 wt%; the DNA content was calculated as 14.3 wt%. The ratio of Ag to DNA was approximately the same before and after the reduction. That is, the Ag nanoparticles consisted largely of Ag, with a low content of impurities such as Na. The TEM images of the Ag nanoparticles showed them to be spherical in shape with an average diameter of <10 nm (Figure 4a). The particle size distribution was determined with AFM (Figure 4b) by using the height profile in the image (Figure 4c). The histogram of the size distribution is shown in Figure 4d; ∼90% of the particles were <10 nm, with a median particle size of 2.6 nm.
Particle images and size distribution of the Ag nanoparticles. (a) Transmission electron microscopy (TEM) and (b) atomic force microscopy (AFM) images of the Ag nanoparticles. (c) Height profile along the indicated line of the AFM image. (d) Histogram of the Ag nanoparticle size distribution as evaluated by AFM. A full color version of this figure is available at Polymer Journal online.
The zeta-potential is a measure of the degree of repulsion between similarly charged particles in a dispersion. A higher zeta-potential indicates a higher stability of the dispersion, which would be more resistant to aggregation. The zeta-potential of the Ag nanoparticles in water was −73.82 mV, which was negatively charged, similar to DNA. The absolute value was higher than that of DNA, −42.17 mV. This result suggested that the surfaces of the Ag nanoparticles were thickly covered with DNA, which led to the high stabilization in aqueous media.
From the ratio of Ag to DNA, a schematic was drawn of the size of the Ag core and the thickness of the outer DNA layer wrapping the core. We applied the literature density value of 1.05 × 104 kg m−3 for Ag,28 and the measured density value of 1.14 × 103 kg m−3 for DNA. For a particle of median size, 2.6 nm in diameter, the Ag core was calculated to be 1.90 nm in diameter and the DNA layer 0.35 nm thick (Figure 5). Adamcik et al.29 reported that the height of single-stranded DNA, dried in air, was 0.35±0.05 nm, according to their AFM observations. Therefore, our results could be explained by supposing that the Ag nanoparticles were coated with a single layer of single-stranded DNA.
In addition, the thermal behavior of the Ag nanoparticles was analyzed by TG. The weight loss of the nanoparticles was low from 250–500 °C, followed by a remarkable change over 500 °C. In contrast, the weight of DNA was continuously decreasing from 250–900 °C (Figure 6a). In the stepwise heating from 200 to 600°C, the weight changed little under 400 °C, and a 15% decrease occurred at 500 °C (Figures 6b and c). This value was identical to the analysis by ICP-AES. In other words, the Ag nanoparticles demonstrated good thermostability up to 400 °C. Moreover, most DNA is expected to decompose by heating over 500 °C, at which point the Ag nanoparticles may show potential as a conductive material.
Long-term stability of the Ag nanoparticles in water
Long-term dispersion stability is necessary for Ag nanoparticle applications. Upon standing for over 6 months, the as-prepared dispersion did not change from a homogeneous dark brown based on visual inspection. The UV–vis absorption spectrum indicated that the particle size and shape were remarkably stable in water (Figure 7). The change in the spectrum was the same or even smaller than that of previously reported highly stable Ag nanoparticles stored at lower concentrations.10, 13, 30 These results demonstrate that the DNA had an important role as a protective agent in addition to serving as the template. This stabilization enabled the DNA-templated Ag nanoparticles to be stored on a large scale and used without re-dispersion.
Conclusions
Widely available DNA from salmon milt was used as a bio-template to prepare Ag nanoparticles. Among the three selected templates, the DNA with the lowest molecular weight (∼20 000 Da, single stranded) was used to prepare the Ag nanoparticle dispersion with the highest Ag concentration (5.3 × 10−2 mol l−1) and without nanoparticle aggregation. Ag accounted for over 85 wt% of these nanoparticles. The nanoparticles were spherical in shape, and their mean diameter was less than 10 nm. The templated DNA in the nanoparticles decomposed at ∼500 °C in air and was mostly removed at higher temperatures. The highly negatively charged Ag nanoparticles demonstrated long-term stability as aqueous dispersions under ambient conditions. Thus, DNA extracted from salmon milt is a viable template for the preparation of stable Ag nanoparticle dispersions. The method presented is inexpensive and simple, highly suitable for mass production and thus would benefit the utility of Ag nanoparticles in the fields of catalysis, optics, electronics and antimicrobial materials.
References
Lee, P. C. & Meisel, D. Adsorption and surface-enhanced raman of dyes on silver and gold sols. J. Phys. Chem. 86, 3391–3395 (1982).
Li, X. L., Zhang, J. H., Xu, W. Q., Jia, H. Y., Wang, X., Yang, B., Zhao, B., Li, B. F. & Ozaki, Y. Mercaptoacetic acid-capped silver nanoparticles colloid: Formation, morphology, and SERS activity. Langmuir. 19, 4285–4290 (2003).
Li, Y. N., Wu, Y. L. & Ong, B. S. Facile synthesis of silver nanoparticles useful for fabrication of high-conductivity elements for printed electronics. J. Am. Chem. Soc. 127, 3266–3267 (2005).
Ahn, B. Y., Duoss, E. B., Motala, M. J., Guo, X. Y., Park, S. I., Xiong, Y. J., Yoon, J., Nuzzo, R. G., Rogers, J. A. & Lewis, J. A. Omnidirectional printing of flexible, stretchable, and spanning silver microelectrodes. Science 323, 1590–1593 (2009).
Chen, D. P., Qiao, X. L., Qiu, X. L. & Chen, J. G. Synthesis and electrical properties of uniform silver nanoparticles for electronic applications. J. Mater. Sci. 44, 1076–1081 (2009).
Ghosh, S. K., Kundu, S., Mandal, M. & Pal, T. Silver and gold nanocluster catalyzed reduction of methylene blue by arsine in a micellar medium. Langmuir. 18, 8756–8760 (2002).
Zheng, L., Zhang, R. C., Ni, Y. X., Du, Q. A., Wang, X., Zhang, J. L. & Li, W. Catalytic performance of Ag nanoparticles templated by polymorphic DNA. Catal. Lett. 139, 145–150 (2010).
Sondi, I. & Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: a case study on E-coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 275, 177–182 (2004).
Rai, M., Yadav, A. & Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 27, 76–83 (2009).
Huang, N. M., Lim, H. N., Radiman, S., Khiew, P. S., Chiu, W. S., Hashim, R. & Chia, C. H. Sucrose ester micellar-mediated synthesis of Ag nanoparticles and the antibacterial properties. Colloid. Surfaces A 353, 69–76 (2010).
Sharma, V. K., Yngard, R. A. & Lin, Y. Silver nanoparticles: Green synthesis and their antimicrobial activities. Adv. Colloid Interface Sci. 145, 83–96 (2009).
Huang, H. Z., Yuan, Q. & Yang, X. R. Preparation and characterization of metal-chitosan nanocomposites. Colloid Surface B 39, 31–37 (2004).
Liu, Y. S., Chen, S. M., Zhong, L. & Wu, G. Z. Preparation of high-stable silver nanoparticle dispersion by using sodium alginate as a stabilizer under gamma radiation. Radiat. Phys. Chem. 78, 251–255 (2009).
Kasyutich, O., Ilari, A., Fiorillo, A., Tatchev, D., Hoell, A. & Ceci, P. Silver ion incorporation and nanoparticle formation inside the cavity of pyrococcus furiosus ferritin: structural and size-distribution analyses. J. Am. Chem. Soc. 132, 3621–3627 (2010).
Petty, J. T., Zheng, J., Hud, N. V. & Dickson, R. M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 126, 5207–5212 (2004).
Molotsky, T., Tamarin, T., Ben Moshe, A., Markovich, G. & Kotlyar, A. B. Synthesis of chiral silver clusters on a DNA template. J. Phys. Chem. C 114, 15951–15954 (2010).
Lan, G. Y., Chen, W. Y. & Chang, H. T. Control of synthesis and optical properties of DNA templated silver nanoclusters by varying DNA length and sequence. RSC Adv 1, 802–807 (2011).
Zhang, X. L., Yu, M., Liu, J. H. & Li, S. M. Fabrication and characterization of Ag nanoparticles based on plasmid DNA as templates. Mater. Lett. 65, 719–721 (2011).
Zinchenko, A. A., Baigl, D. M., Chen, N., Pyshkina, O., Endo, K., Sergeyev, V. G. & Yoshikawa, K. Conformational behavior of giant DNA through binding with Ag+ and metallization. Biomacromolecules 9, 1981–1987 (2008).
Yamada, M., Yokota, M., Kaya, M., Satoh, S., Jonganurakkun, B., Nomizu, M. & Nishi, N. Preparation of novel bio-matrix by the complexation of DNA and metal ions. Polymer 46, 10102–10112 (2005).
Iwata, K., Sawadaishi, T., Nishimura, S., Tokura, S. & Nishi, N. Utilization of DNA as functional materials: Preparation of filters containing DNA insolubilized with alginic acid gel. Int. J. Biol. Macromol. 18, 149–150 (1996).
Yamada, M., Kato, K., Nomizu, M., Sakairi, N., Ohkawa, K., Yamamoto, H. & Nishi, N. Preparation and characterization of DNA films induced by UV irradiation. Chem. Eur. J 8, 1407–1412 (2002).
Satoh, S., Fugetsu, B., Nomizu, M. & Nishi, N. Functional DNA-silica composite prepared by sol-gel method. Polym. J. 37, 94–101 (2005).
Yamane, T. & Davidson, N. On complexing of deoxyribonucleic acid by silver(I). Biochim. Biophys. Acta 55, 609–621 (1962).
Izatt, R. M., Christen, J. J. & Rytting, J. H. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem. Rev. 71, 439–481 (1971).
Huang, H. H., Ni, X. P., Loy, G. L., Chew, C. H., Tan, K. L., Loh, F. C., Deng., J. F. & Xu, G. Q. Photochemical formation of silver nanoparticles in poly(N-vinylpyrrolidone). Langmuir. 12, 909–912 (1996).
Fornasiero, D. & Grieser, F. The kinetics of electrolyte induced aggregation of Carey Lea Silver colloids. J. Colloid Interface Sci. 141, 168–179 (1991).
The Chemical Society of Japan. Kagaku Binran Kiso-hen I 5th edn 32, ((Maruzen, Tokyo, Japan, 2004) in Japanese.
Adamcik, J., Klinov, D. V., Witz, G., Sekatskii, S. K. & Dietler, G. Observation of single-stranded DNA on mica and highly oriented pyrolytic graphite by atomic force microscopy. FEBS Lett. 580, 5671–5675 (2006).
Liu, J. Y. & Hurt, R. H. Ion release kinetics and particle persistence in aqueous nano-silver colloids. Environ. Sci. Technol. 44, 2169–2175 (2010).
Acknowledgements
The analysis of the Ag nanoparticles was conducted using STEM, ICP-AES, and a particle analyzer at the OPEN FACILITY, Sousei Hall, Hokkaido University, and TEM at the Center for Advanced Research of Energy and Materials, Faculty of Engineering, Hokkaido University. We would like to thank Dr Masahito Sugi and Mr. Masao Nishihara, Nissei Bio Co., Ltd., for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Takeshima, T., Sun, L., Wang, Y. et al. Salmon milt DNA as a template for the mass production of Ag nanoparticles. Polym J 46, 36–41 (2014). https://doi.org/10.1038/pj.2013.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pj.2013.58
Keywords
This article is cited by
-
Small DNA additives to polyelectrolyte multilayers promote formation of ultrafine gold nanoparticles with enhanced catalytic activity
Colloid and Polymer Science (2019)
-
Reusable resistive aptasensor for Pb(II) based on the Pb(II)-induced despiralization of a DNA duplex and formation of a G-quadruplex
Microchimica Acta (2018)