Abstract
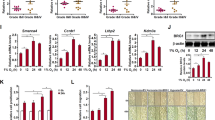
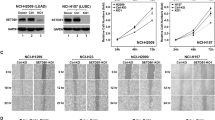
Epigenetic regulators are attractive targets for the development of new cancer therapies. Among them, the ATP-dependent chromatin remodeling complexes control the chromatin architecture and have important roles in gene regulation. They are often found to be mutated and de-regulated in cancers, but how they influence the cancer gene expression program during cancer initiation and progression is not fully understood. Here we show that the INO80 chromatin remodeling complex is required for oncogenic transcription and tumor growth in non-small-cell lung cancer (NSCLC). Ino80, the SWI/SNF ATPase in the complex, is highly expressed in NSCLC cells compared with normal lung epithelia cells. Further, its expression, as well as that of another subunit Ino80B, negatively correlates with disease prognosis in lung cancer patients. Functionally, INO80 silencing inhibits NSCLC cell proliferation and anchorage-independent growth in vitro and tumor formation in mouse xenografts. It occupies enhancer regions near lung cancer-associated genes, and its occupancy correlates with increased genome accessibility and enhanced expression of downstream genes. Together, our study defines a critical role of INO80 in promoting oncogenic transcription and NSCLC tumorigenesis, and reveals a potential treatment strategy for inhibiting the cancer transcription network by targeting the INO80 chromatin remodeling complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bender E . Epidemiology: the dominant malignancy. Nature 2014; 513: S2–S3.
Lennon FE, Cianci GC, Cipriani NA, Hensing TA, Zhang HJ, Chen CT et al. Lung cancer-a fractal viewpoint. Nat Rev Clin Oncol 2015; 12: 664–675.
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK . Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014; 14: 535–546.
Reck M, Heigener DF, Mok T, Soria JC, Rabe KF . Management of non-small-cell lung cancer: recent developments. Lancet 2013; 382: 709–719.
Viktorsson K, Lewensohn R, Zhivotovsky B . Systems biology approaches to develop innovative strategies for lung cancer therapy. Cell Death Dis 2014; 5: e1260.
Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M . Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012; 11: 384–400.
Belinsky SA . Unmasking the lung cancer epigenome. Annu Rev Physiol 2015; 77: 453–474.
Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE . Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol 2013; 3: 261.
Liu SV, Fabbri M, Gitlitz BJ, Laird-Offringa IA . Epigenetic therapy in lung cancer. Front Oncol 2013; 3: 135.
Hargreaves DC, Crabtree GR . ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21: 396–420.
Narlikar GJ, Sundaramoorthy R, Owen-Hughes T . Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013; 154: 490–503.
Helming KC, Wang X, Roberts CW . Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 2014; 26: 309–317.
Kadoch C, Crabtree GR . Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 2015; 1: e1500447.
Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O . SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 2015; 10: 145–171.
Conaway RC, Conaway JW . The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci 2009; 34: 71–77.
Morrison AJ, Shen X . Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 2009; 10: 373–384.
Watanabe S, Peterson CL . The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb Symp Quant Biol 2010; 75: 35–42.
Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 2014; 14: 575–591.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Kim J, Orkin SH . Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med 2011; 3: 75.
Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY . Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008; 2: 333–344.
Stewart DJ . Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 2014; 106: djt356.
Arenberg DA, Keane MP, DiGiovine B, Kunkel SL, Morris SB, Xue YY et al. Epithelial-neutrophil activating peptide (ENA-78) is an important angiogenic factor in non-small cell lung cancer. J Clin invest 1998; 102: 465–472.
Kolluri SK, Bruey-Sedano N, Cao X, Lin B, Lin F, Han YH et al. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol Cell Biol 2003; 23: 8651–8667.
Linxweiler M, Linxweiler J, Barth M, Benedix J, Jung V, Kim YJ et al. Sec62 bridges the gap from 3q amplification to molecular cell biology in non-small cell lung cancer. Am J Pathol 2012; 180: 473–483.
Baykara O, Bakir B, Buyru N, Kaynak K, Dalay N . Amplification of chromosome 8 genes in lung cancer. J Cancer 2015; 6: 270–275.
Kramer A, Green J, Pollard J Jr, Tugendreich S . Causal analysis approaches in Ingenuity pathway analysis. Bioinformatics 2014; 30: 523–530.
El-Telbany A, Ma PC . Cancer genes in lung cancer: racial disparities: are there any? Genes Cancer 2012; 3: 467–480.
Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P et al. Genomic aberrations in lung adenocarcinoma in never smokers. PloS One 2010; 5: e15145.
Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL . Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011; 144: 200–213.
Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL . A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 2013; 340: 195–199.
Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M . Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 2011; 12: 36–47.
Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 2011; 21: 1650–1658.
Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife 2015; 4 doi:10.7554/eLife.06857.
Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S . The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res 2013; 23: 1396–1413.
Han L, Diao L, Yu S, Xu X, Li J, Zhang R et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 2015; 28: 515–528.
Acknowledgements
We thank the NIEHS Animal, Epigenomics and Bioinformatics core facilities for assistance with various techniques and experiments. This study was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Z01ES102745 (to GH) and by National Institutes of Health Grants CA042978 and CA161494 (to ADC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, S., Zhou, B., Wang, L. et al. INO80 is required for oncogenic transcription and tumor growth in non-small cell lung cancer. Oncogene 36, 1430–1439 (2017). https://doi.org/10.1038/onc.2016.311
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.311
This article is cited by
-
Titanium(IV) immobilized affinity chromatography facilitated phosphoproteomics analysis of salivary extracellular vesicles for lung cancer
Analytical and Bioanalytical Chemistry (2022)
-
The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling
Nature Communications (2018)
-
Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity
Cellular and Molecular Life Sciences (2018)