Abstract
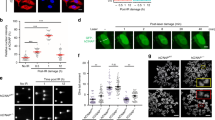
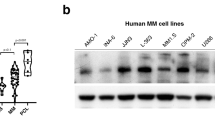
MMSET/WHSC1 is a histone methyltransferase (HMT) overexpressed in t(4;14)+ multiple myeloma (MM) patients, believed to be the driving factor in the pathogenesis of this MM subtype. MMSET overexpression in MM leads to an increase in histone 3 lysine 36 dimethylation (H3K36me2), and a decrease in histone 3 lysine 27 trimethylation (H3K27me3), as well as changes in proliferation, gene expression and chromatin accessibility. Prior work linked methylation of histones to the ability of cells to undergo DNA damage repair. In addition, t(4;14)+ patients frequently relapse after regimens that include DNA damage-inducing agents, suggesting that MMSET may play a role in DNA damage repair and response. In U2OS cells, we found that MMSET is required for efficient non-homologous end joining as well as homologous recombination. Loss of MMSET led to loss of expression of several DNA repair proteins, as well as decreased recruitment of DNA repair proteins to sites of DNA double-strand breaks (DSBs). By using genetically matched MM cell lines that had either high (pathological) or low (physiological) expression of MMSET, we found that MMSET-high cells had increased damage at baseline. Upon addition of a DNA-damaging agent, MMSET-high cells repaired DNA damage at an enhanced rate and continued to proliferate, whereas MMSET-low cells accumulated DNA damage and entered cell cycle arrest. In a murine xenograft model using t(4;14)+ KMS11 MM cells harboring an inducible MMSET shRNA, depletion of MMSET enhanced the efficacy of chemotherapy, inhibiting tumor growth and extending survival. These findings help explain the poorer prognosis of t(4;14) MM and further validate MMSET as a potential therapeutic target in MM and other cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Bergsagel PL, Kuehl WM . Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 2005; 23: 6333–6338.
Palumbo A, Anderson K . Multiple myeloma. N Engl J Med 2011; 364: 1046–1060.
Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL . The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998; 92: 3025–3034.
Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 2003; 101: 1520–1529.
Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 1998; 7: 1071–1082.
Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiaer E, Rossing HH, Johansen JV et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res 2011; 17: 2919–2933.
Hudlebusch HR, Skotte J, Santoni-Rugiu E, Zimling ZG, Lees MJ, Simon R et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res 2011; 71: 4226–4235.
Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011; 117: 211–220.
Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci USA 2012; 109: 13549–13554.
Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Ezponda T, Shah MY et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet 2014; 10: e1004566.
Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood 2008; 111: 856–864.
Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 2011; 44: 609–620.
Gourzones-Dmitriev C, Kassambara A, Sahota S, Reme T, Moreaux J, Bourquard P et al. DNA repair pathways in human multiple myeloma: role in oncogenesis and potential targets for treatment. Cell Cycle 2013; 12: 2760–2773.
Palumbo A, Cavallo F, Gay F, Di Raimondo F, Ben Yehuda D, Petrucci MT et al. Autologous transplantation and maintenance therapy in multiple myeloma. N Engl J Med 2014; 371: 895–905.
Jaksic W, Trudel S, Chang H, Trieu Y, Qi X, Mikhael J et al. Clinical outcomes in t(4;14) multiple myeloma: a chemotherapy-sensitive disease characterized by rapid relapse and alkylating agent resistance. J Clin Oncol 2005; 23: 7069–7073.
Roussel M, Moreau P, Huynh A, Mary JY, Danho C, Caillot D et al. Bortezomib and high-dose melphalan as conditioning regimen before autologous stem cell transplantation in patients with de novo multiple myeloma: a phase 2 study of the Intergroupe Francophone du Myelome (IFM). Blood 2010; 115: 32–37.
Cook G, Williams C, Brown JM, Cairns DA, Cavenagh J, Snowden JA et al. High-dose chemotherapy plus autologous stem-cell transplantation as consolidation therapy in patients with relapsed multiple myeloma after previous autologous stem-cell transplantation (NCRI Myeloma X Relapse [Intensive trial]): a randomised, open-label, phase 3 trial. Lancet Oncol 2014; 15: 874–885.
Hajdu I, Ciccia A, Lewis SM, Elledge SJ . Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci USA 2011; 108: 13130–13134.
Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011; 470: 124–128.
Pei H, Wu X, Liu T, Yu K, Jelinek DF, Lou Z . The histone methyltransferase MMSET regulates class switch recombination. J Immunol 2013; 190: 756–763.
Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA 2011; 108: 540–545.
Jha DK, Strahl BD . An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun 2014; 5: 3965.
Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 2014; 5: 4091.
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 2014; 21: 366–374.
Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 2014; 3: e02482.
Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014; 7: 2006–2018.
Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010; 17: 1144–1151.
Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 2013; 32: 2882–2890.
Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 2010; 29: 1446–1457.
Lans H, Marteijn JA, Vermeulen W . ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 2012; 5: 4.
Panier S, Boulton SJ . Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014; 15: 7–18.
Li L, Robert C, Rassool FV The role of error-prone alternative non-homologous end-joining in genomic instability in cancer. In: Vengrova S (ed). DNA Repair and Human Health. InTech: Rijeka, Croatia, 2011.
Povirk LF, Han YH, Steighner RJ . Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5' extensions. Biochemistry 1989; 28: 5808–5814.
Lupi M, Cappella P, Matera G, Natoli C, Ubezio P . Interpreting cell cycle effects of drugs: the case of melphalan. Cancer Chemother Pharmacol 2006; 57: 443–457.
Liao W, McNutt MA, Zhu WG . The comet assay: a sensitive method for detecting DNA damage in individual cells. Methods 2009; 48: 46–53.
Chambliss AB, Wu PH, Chen WC, Sun SX, Wirtz D . Simultaneously defining cell phenotypes, cell cycle, and chromatin modifications at single-cell resolution. FASEB J 2013; 27: 2667–2676.
Guo Q, Phillip JM, Majumdar S, Wu PH, Chen J, Calderon-Colon X et al. Modulation of keratocyte phenotype by collagen fibril nanoarchitecture in membranes for corneal repair. Biomaterials 2013; 34: 9365–9372.
Chen WC, Wu PH, Phillip JM, Khatau SB, Choi JM, Dallas MR et al. Functional interplay between the cell cycle and cell phenotypes. Integr Biol 2013; 5: 523–534.
Sulli G, Di Micco R, d'Adda di Fagagna F . Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 2012; 12: 709–720.
Ransom M, Dennehey BK, Tyler JK . Chaperoning histones during DNA replication and repair. Cell 2010; 140: 183–195.
Soria G, Polo SE, Almouzni G . Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 2012; 46: 722–734.
Berkovich E, Monnat RJ Jr, Kastan MB . Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 2007; 9: 683–690.
Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB . Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci USA 2013; 110: 16874–16879.
Deininger P . Alu elements: know the SINEs. Genome Biol 2011; 12: 236.
Englander EW, Howard BH . Nucleosome positioning by human Alu elements in chromatin. J Biol Chem 1995; 270: 10091–10096.
Englander EW, Wolffe AP, Howard BH . Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J Biol Chem 1993; 268: 19565–19573.
Englander EW, Howard BH . Alu-mediated detection of DNA damage in the human genome. Mutat Res 1997; 385: 31–39.
Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011; 332: 1443–1446.
Min DJ, Ezponda T, Kim MK, Will CM, Martinez-Garcia E, Popovic R et al. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia 2013; 27: 686–694.
Cann KL, Dellaire G . Heterochromatin and the DNA damage response: the need to relax. Biochem Cell Biol 2011; 89: 45–60.
Wang Q, Goldstein M . Small RNAs recruit chromatin modifying enzymes MMSET and Tip60 to reconfigure damaged DNA upon double-strain break and facilitate repair. Cancer Res 2016 [Epub ahead of print].
Hsiao KY, Mizzen CA . Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol 2013; 5: 157–165.
Tuzon CT, Spektor T, Kong X, Congdon LM, Wu S, Schotta G et al. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep 2014; 8: 430–438.
Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ 3rd et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 2009; 284: 34283–34295.
Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T . Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006; 10: 105–116.
Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008; 2: 437–447.
Banath JP, Banuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL . Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 2009; 315: 1505–1520.
Gaymes TJ, Padua RA, Pla M, Orr S, Omidvar N, Chomienne C et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res 2006; 4: 563–573.
Lee JH, Choy ML, Ngo L, Foster SS, Marks PA . Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA 2010; 107: 14639–14644.
Sousa MM, Zub KA, Aas PA, Hanssen-Bauer A, Demirovic A, Sarno A et al. An inverse switch in DNA base excision and strand break repair contributes to melphalan resistance in multiple myeloma cells. PLoS One 2013; 8: e55493.
Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev 2010; 24: 837–852.
Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB et al. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res 2002; 8: 3008–3018.
Acknowledgements
This study was supported by NIH grant R01CA123204, Leukemia & Lymphoma Society Specialized Center for Research Award, Physical Sciences Oncology Center U54CA143869, Celgene and the Samuel Waxman Cancer Research Foundation (to JDL); NIH grants R01CA174388 and U54CA143868 (to DW); and Canadian Institute of Health and Research operating grant (287300; to NJB). We thank Dr Gaëlle Legube for the AsiSI-U2OS cells and members of the Licht lab for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
JDL receives funding from Celgene.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Shah, M., Martinez-Garcia, E., Phillip, J. et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 35, 5905–5915 (2016). https://doi.org/10.1038/onc.2016.116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.116
This article is cited by
-
Association of WHSC1/NSD2 and T-cell infiltration with prostate cancer metastasis and prognosis
Scientific Reports (2023)
-
A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization
Nature Chemical Biology (2022)
-
Classification models for Invasive Ductal Carcinoma Progression, based on gene expression data-trained supervised machine learning
Scientific Reports (2020)
-
H3K36 dimethylation by MMSET promotes classical non-homologous end-joining at unprotected telomeres
Oncogene (2020)
-
Proteosomal degradation of NSD2 by BRCA1 promotes leukemia cell differentiation
Communications Biology (2020)