Abstract
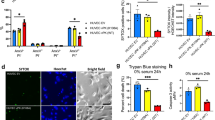
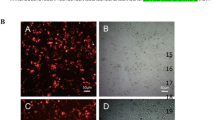
The tumor suppressor p53 is a central regulatory molecule of apoptosis and is commonly mutated in tumors. Kaposi’s sarcoma-associated herpesvirus (KSHV)-related malignancies express wild-type p53. Accordingly, KSHV encodes proteins that counteract the cell death-inducing effects of p53. Here, the effects of all KSHV genes on the p53 signaling pathway were systematically analyzed using the reversely transfected cell microarray technology. With this approach we detected eight KSHV-encoded genes with potent p53 inhibiting activity in addition to the previously described inhibitory effects of KSHV genes ORF50, K10 and K10.5. Interestingly, the three most potent newly identified inhibitors were KSHV structural proteins, namely ORF22 (glycoprotein H), ORF25 (major capsid protein) and ORF64 (tegument protein). Validation of these results with a classical transfection approach showed that these proteins inhibited p53 signaling in a dose-dependent manner and that this effect could be reversed by small interfering RNA-mediated knockdown of the respective viral gene. All three genes inhibited p53-mediated apoptosis in response to Nutlin-3 treatment in non-infected and KSHV-infected cells. Addressing putative mechanisms, we could show that these proteins could also inhibit the transactivation of the promoters of apoptotic mediators of p53 such as BAX and PIG3. Altogether, we demonstrate for the first time that structural proteins of KSHV can counteract p53-induced apoptosis. These proteins are expressed in the late lytic phase of the viral life cycle and are incorporated into the KSHV virion. Accordingly, these genes may inhibit cell death in the productive and in the early entrance phase of KSHV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994; 266: 1865–1869.
Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM . Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 1995; 332: 1186–1191.
Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995; 86: 1276–1280.
Dittmer DP . Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer Res 2003; 63: 2010–2015.
Dittmer DP . Restricted Kaposi’s sarcoma (KS) herpesvirus transcription in KS lesions from patients on successful antiretroviral therapy. MBio 2011; 2: e00138–11.
Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A et al. Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J Virol 1999; 73: 2232–2242.
Schulz TF, Chang Y . KSHV gene expression and regulation. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, et al (eds). Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis Chapter 28 Cambridge University Press, Cambridge, 2007.
Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 1997; 71: 715–719.
Stürzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E et al. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi’s sarcoma. Int J Cancer 1997; 72: 68–71.
Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer NH et al. Monocytes in Kaposi’s sarcoma lesions are productively infected by human herpesvirus 8. J Virol 1997; 71: 7963–7968.
Piette J, Neel H, Marechal V . Mdm2: keeping p53 under control. Oncogene 1997; 15: 1001–1010.
Hollstein M, Sidransky D, Vogelstein B, Harris CC . p53 mutations in human cancers. Science 1991; 253: 49–53.
Lane DP . Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16.
Levine AJ, Momand J, Finlay CA . The p53 tumour suppressor gene. Nature 1991; 351: 453–456.
Petre CE, Sin SH, Dittmer DP . Functional p53 signaling in Kaposi’s sarcoma-associated herpesvirus lymphomas: implications for therapy. J Virol 2007; 81: 1912–1922.
Tornesello ML, Biryahwaho B, Downing R, Hatzakis A, Alessi E, Cusini M et al. TP53 codon 72 polymorphism in classic, endemic and epidemic Kaposi’s sarcoma in African and Caucasian patients. Oncology 2009; 77: 328–334.
Friborg J Jr, Kong W, Hottiger MO, Nabel GJ . p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 1999; 402: 889–894.
Gwack Y, Hwang S, Byun H, Lim C, Kim JW, Choi EJ et al. Kaposi’s sarcoma-associated herpesvirus open reading frame 50 represses p53-induced transcriptional activity and apoptosis. J Virol 2001; 75: 6245–6248.
Lee HR, Toth Z, Shin YC, Lee JS, Chang H, Gu W et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J Virol 2009; 83: 6739–6747.
Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y . Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol 2001; 75: 429–438.
Chen W, Hilton IB, Staudt MR, Burd CE, Dittmer DP . Distinct p53, p53:LANA, and LANA complexes in Kaposi’s sarcoma—associated herpesvirus lymphomas. J Virol 2010; 84: 3898–3908.
Sarek G, Kurki S, Enback J, Iotzova G, Haas J, Laakkonen P et al. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest 2007; 117: 1019–1028.
Ye F, Lattif AA, Xie J, Weinberg A, Gao S . Nutlin-3 induces apoptosis, disrupts viral latency and inhibits expression of angiopoietin-2 in Kaposi sarcoma tumor cells. Cell Cycle 2012; 11: 1393–1399.
Vassilev LT . Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle 2004; 3: 419–421.
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004; 303: 844–848.
Sarek G, Ma L, Enback J, Jarviluoma A, Moreau P, Haas J et al. Kaposi’s sarcoma herpesvirus lytic replication compromises apoptotic response to p53 reactivation in virus-induced lymphomas. Oncogene 2013; 32: 1091–1098.
Sander G, Konrad A, Thurau M, Wies E, Leubert R, Kremmer E et al. Intracellular localization map of human herpesvirus 8 proteins. J Virol 2008; 82: 1908–1922.
Konrad A, Wies E, Thurau M, Marquardt G, Naschberger E, Hentschel S et al. A systems biology approach to identify the combination effects of human herpesvirus 8 genes on NF-kappaB activation. J Virol 2009; 83: 2563–2574.
Kuhn E, Naschberger E, Konrad A, Croner RS, Britzen-Laurent N, Jochmann R et al. A novel chip-based parallel transfection assay to evaluate paracrine cell interactions. Lab Chip 2012; 12: 1363–1372.
Ziauddin J, Sabatini DM . Microarrays of cells expressing defined cDNAs. Nature 2001; 411: 107–110.
Graham FL, Smiley J, Russell WC, Nairn R . Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 1977; 36: 59–74.
Moran E . Interaction of adenoviral proteins with pRB and p53. FASEB J 1993; 7: 880–885.
Naranatt PP, Akula SM, Chandran B . Characterization of gamma2-human herpesvirus-8 glycoproteins gH and gL. Arch Virol 2002; 147: 1349–1370.
Hahn A, Birkmann A, Wies E, Dorer D, Mahr K, Stürzl M et al. Kaposi’s sarcoma-associated herpesvirus gH/gL: glycoprotein export and interaction with cellular receptors. J Virol 2009; 83: 396–407.
Hahn AS, Kaufmann JK, Wies E, Naschberger E, Panteleev-Ivlev J, Schmidt K et al. The ephrin receptor tyrosine kinase A2 is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus. Nat Med 2012; 18: 961–966.
Myoung J, Ganem D . Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 2011; 174: 12–21.
Miyashita T, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B . A model for p53-induced apoptosis. Nature 1997; 389: 300–305.
Barak Y, Juven T, Haffner R, Oren M . mdm2 expression is induced by wild type p53 activity. EMBO J 1993; 12: 461–468.
Stürzl M, Konrad A, Sander G, Wies E, Neipel F, Naschberger E et al. High throughput screening of gene functions in mammalian cells using reversely transfected cell arrays: review and protocol. Comb Chem High Throughput Screen 2008; 11: 159–172.
Nakamura H, Li M, Zarycki J, Jung JU . Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 2001; 75: 7572–7582.
Leidal AM, Cyr DP, Hill RJ, Lee PW, McCormick C . Subversion of autophagy by Kaposi’s sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe 2012; 11: 167–180.
Pertel PE . Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J Virol 2002; 76: 4390–4400.
Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K et al. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol 1992; 66: 2240–2250.
Spaete RR, Perot K, Scott PI, Nelson JA, Stinski MF, Pachl C . Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in transport of gH to the cell surface. Virology 1993; 193: 853–861.
Jerome KR, Chen Z, Lang R, Torres MR, Hofmeister J, Smith S et al. HSV and glycoprotein J inhibit caspase activation and apoptosis induced by granzyme B or Fas. J Immunol 2001; 167: 3928–3935.
Nakamichi K, Kuroki D, Matsumoto Y, Otsuka H . Bovine herpesvirus 1 glycoprotein G is required for prevention of apoptosis and efficient viral growth in rabbit kidney cells. Virology 2001; 279: 488–498.
Nealon K, Newcomb WW, Pray TR, Craik CS, Brown JC, Kedes DH . Lytic replication of Kaposi’s sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J Virol 2001; 75: 2866–2878.
Jochmann R, Pfannstiel J, Chudasama P, Kuhn E, Konrad A, Stürzl M . O-GlcNAc transferase inhibits KSHV propagation and modifies replication relevant viral proteins as detected by systematic O-GlcNAcylation analysis. Glycobiology 2013; 23: 1114–1130.
Ilkow CS, Goping IS, Hobman TC . The Rubella virus capsid is an anti-apoptotic protein that attenuates the pore-forming ability of Bax. PLoS Pathog 2011; 7: e1001291.
Urbanowski MD, Hobman TC . The West Nile virus capsid protein blocks apoptosis through a phosphatidylinositol 3-kinase-dependent mechanism. J Virol 2013; 87: 872–881.
Kwon JA, Rho HM . Transcriptional repression of the human p53 gene by hepatitis B viral core protein (HBc) in human liver cells. Biol Chem 2003; 384: 203–212.
Rozen R, Sathish N, Li Y, Yuan Y . Virion-wide protein interactions of Kaposi’s sarcoma-associated herpesvirus. J Virol 2008; 82: 4742–4750.
Sathish N, Wang X, Yuan Y . Tegument proteins of Kaposi’s sarcoma-associated herpesvirus and related gamma-herpesviruses. Front Microbiol 2012; 3: 98.
Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K et al. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol 2011; 85: 10899–10904.
Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003; 424: 516–523.
Gross A, Jockel J, Wei MC, Korsmeyer SJ . Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J 1998; 17: 3878–3885.
Kurosu T, Wu N, Oshikawa G, Kagechika H, Miura O . Enhancement of imatinib-induced apoptosis of BCR/ABL-expressing cells by nutlin-3 through synergistic activation of the mitochondrial apoptotic pathway. Apoptosis 2010; 15: 608–620.
Voltan R, Secchiero P, Corallini F, Zauli G . Selective induction of TP53I3/p53-inducible gene 3 (PIG3) in myeloid leukemic cells, but not in normal cells, by Nutlin-3. Mol Carcinog (e-pub ahead of print 28 November 2012; doi: 10.1002/mc.21985).
Grundhoff A, Ganem D . Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest 2004; 113: 124–136.
Austgen K, Oakes SA, Ganem D . Multiple defects, including premature apoptosis, prevent Kaposi’s sarcoma-associated herpesvirus replication in murine cells. J Virol 2012; 86: 1877–1882.
Lagunoff M, Carroll PA . Inhibition of apoptosis by the gamma-herpesviruses. Int Rev Immunol 2003; 22: 373–399.
Moore PS . KSHV manipulation of the cell cycle and apoptosis. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, et al. (eds). Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis Chapter 30 Cambridge University Press, Cambridge, 2007.
Hanon E, Meyer G, Vanderplasschen A, Dessy-Doize C, Thiry E, Pastoret PP . Attachment but not penetration of bovine herpesvirus 1 is necessary to induce apoptosis in target cells. J Virol 1998; 72: 7638–7641.
Morris SJ, Price GE, Barnett JM, Hiscox SA, Smith H, Sweet C . Role of neuraminidase in influenza virus-induced apoptosis. J Gen Virol 1999; 80 (Pt 1): 137–146.
Raftery MJ, Behrens CK, Muller A, Krammer PH, Walczak H, Schonrich G . Herpes simplex virus type 1 infection of activated cytotoxic T cells: induction of fratricide as a mechanism of viral immune evasion. J Exp Med 1999; 190: 1103–1114.
de Oliveira DE, Ballon G, Cesarman E . NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol 2010; 18: 248–257.
Chandran B . Early events in Kaposi’s sarcoma-associated herpesvirus infection of target cells. J Virol 2009; 84: 2188–2199.
Burbelo PD, Issa AT, Ching KH, Wyvill KM, Little RF, Iadarola MJ et al. Distinct profiles of antibodies to Kaposi sarcoma-associated herpesvirus antigens in patients with Kaposi sarcoma, multicentric Castleman disease, and primary effusion lymphoma. J Infect Dis 2010; 201: 1919–1922.
Katano H, Sato Y, Kurata T, Mori S, Sata T . Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi’s sarcoma, and multicentric Castleman’s disease. Virology 2000; 269: 335–344.
Marcelin AG, Motol J, Guihot A, Caumes E, Viard JP, Dussaix E et al. Relationship between the quantity of Kaposi sarcoma-associated herpesvirus (KSHV) in peripheral blood and effusion fluid samples and KSHV-associated disease. J Infect Dis 2007; 196: 1163–1166.
Stürzl M, Gaus D, Dirks WG, Ganem D, Jochmann R . Kaposi’s sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int J Cancer 2013; 132: 1954–1958.
Konrad A, Jochmann R, Kuhn E, Naschberger E, Chudasama P, Stürzl M . Reverse transfected cell microarrays in infectious disease research. Methods Mol Biol 2011; 706: 107–118.
Stürzl M, Konrad A, Alkharsah KR, Jochmann R, Thurau M, Marquardt G et al. The contribution of systems biology and reverse genetics to the understanding of Kaposi’s sarcoma-associated herpesvirus pathogenesis in endothelial cells. Thromb Haemost 2009; 102: 1117–1134.
Naschberger E, Werner T, Vicente AB, Guenzi E, Topolt K, Leubert R et al. Nuclear factor-kappaB motif and interferon-alpha-stimulated response element co-operate in the activation of guanylate-binding protein-1 expression by inflammatory cytokines in endothelial cells. Biochem J 2004; 379 (Pt 2): 409–420.
Britzen-Laurent N, Lipnik K, Ocker M, Naschberger E, Schellerer VS, Croner RS et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis 2012; 34: 153–162.
Acknowledgements
We thank Professor Moshe Oren, Professor Wafik S El-Deiry and Professor Bert Vogelstein for sharing plasmids. In addition, we thank PD Dr Reiner Strick and PD Dr Pamela Strissel (University Medical Center Erlangen, Department of Obstetrics and Gynecology) for providing the MCF7 cells. This work was supported by grants of the German Federal Ministry of Education and Research (BMBF, Polyprobe-Study), the Deutsche Forschungsgemeinschaft (DFG-GRK1071, STU238/6-1, SFB796 (sub-project B9)) and the German Cancer Aid (109510). Additional support was obtained from the Interdisciplinary Center for Clinical Research (IZKF) and the Emerging Fields Initiative of the Friedrich-Alexander University of Erlangen to MS, by a grant for the promotion of young researchers (ELAN) of the University Medical Center Erlangen to AK and a grant of the ‘Programm zur Förderung der Chancengleichheit für Frauen in Forschung und Lehre (FFL)’ to PC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Chudasama, P., Konrad, A., Jochmann, R. et al. Structural proteins of Kaposi’s sarcoma-associated herpesvirus antagonize p53-mediated apoptosis. Oncogene 34, 639–649 (2015). https://doi.org/10.1038/onc.2013.595
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.595
Keywords
This article is cited by
-
Pathogenesis of Human Gammaherpesviruses: Recent Advances
Current Clinical Microbiology Reports (2019)
-
A role for MALT1 activity in Kaposi’s sarcoma-associated herpes virus latency and growth of primary effusion lymphoma
Leukemia (2017)