Abstract
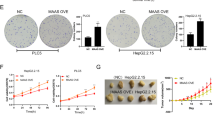
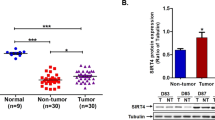
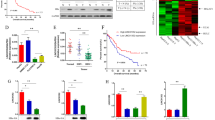
Hepatocellular carcinoma (HCC) is one of the most deadly cancers. Aberrant oncogenic activation of the Wnt/β-catenin signaling pathway contributes to hepatocellular carcinogenesis. Various epigenetic modifications of the Wnt antagonist secreted frizzled-related protein (SFRP) family have been implicated in regulating Wnt signaling. Here, we report that Hepatitis C virus (HCV) core protein downregulates SFRP1 expression when it is expressed in Huh7 and HepG2 cells. SFRP1 expression can be effectively restored by using either a DNA methylation inhibitor alone or in combination with a histone deacetylase inhibitor. DNA methylation analysis of the SFRP1 promoter revealed that cytosine-phosphate-guanine (CpG) islands close to the transcriptional start site (TSS) in the SFRP1 promoter were hypermethylated in core-expressing Huh7 cells, suggesting that HCV core protein may downregulate SFRP1 expression by inducing hypermethylation of the SFRP1 promoter. Chromatin immunoprecipitation revealed that HCV core protein markedly increased the expression level and binding of DNA methyltransferase-1 (Dnmt1) and histone deacetylase-1 (HDAC1) to the TSS of the SFRP1 promoter region, resulting in repression of acetyl-histone H3-binding capacity to SFRP1 promoter and the eventual epigenetic silencing of SFRP1 expression. Furthermore, the core protein-promoted cell proliferation, migration and invasiveness were effectively abrogated either by Dnmt1 knockdown or restoration of SFRP1 expression in hepatoma cells. Dnmt1 knockdown or SFRP1 overexpression also inhibited HCV core-induced epithelial–mesenchymal transition (EMT) and significantly decreased the expression levels of activated β-catenin and Wnt/β-catenin target genes, c-Myc and cyclin D1. We further showed that knockdown of Dnmt1 and restoration of SFRP1 inhibited core-induced in vivo tumor growth and aggressiveness in a xenograft HCC model. Taken together, our results strongly suggest that the HCV core-induced epigenetic silencing of SFRP1 may lead to the activation of the Wnt signaling pathway and thus contribute to HCC aggressiveness through induction of EMT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kaur P, Mani S, Cros MP, Scoazec JY, Chemin I, Hainaut P et al. Epigenetic silencing of sFRP1 activates the canonical Wnt pathway and contributes to increased cell growth and proliferation in hepatocellular carcinoma. Tumour Biol 2012; 33: 325–336.
Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC et al. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer 2007; 121: 1028–1035.
Clément S, Pascarella S, Conzelmann S, Gonelle-Gispert C, Guilloux K, Negro F . The hepatitis C Core protein indirectly induces alpha-smooth muscle actin expression in hepatic stellate cells via interleukin-8. J Hepatol 2010; 52: 635–643.
Kato N, Yoshida H, Ono-Nita SK, Kato J, Goto T, Otsuka M et al. Activation of intracellular signaling by hepatitis B and C viruses:C-viral core is the most potent signal inducer. Hepatology 2000; 32: 405–412.
Guo N, Cheng D, Li ZH, Zhou QB, Zhou JJ, Lin Q et al. Transfection of HCVc mproves hTERT expression through STAT3 pathway by epigenetic regulation in Huh7 cells. J Cell Biochem 2012; 113: 3419–3426.
Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP et al. Wnt signaling through inhiition of β-catenin degration in an intact Axin1 complex. Cell 2012; 149: 1245–1256.
Clevers H, Nusse R . Wnt/β-catenin signaling and disease. Cell 2012; 149: 1192–1205.
Austinat M, Dunsch R, Wittekind C, Tannapfel A, Gebhardt R, Gaunitz F . Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer 2008; 7: 21.
Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 2008; 13: 529–541.
Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004; 36: 417–422.
Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F et al. Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One 2011; 6: e27496.
Bi Y, Huang J, He Y, Zhu GH, Su Y, He BC et al. Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J Cell Biochem 2009; 108: 295–303.
Huang J, Bi Y, Zhu GH, He Y, Su Y, He BC et al. Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells. Liver Int 2009; 29: 1569–1581.
Danam RP, Howell SR, Brent TP, Harris LC . Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histoneacetylation and methyl-CpG binding proteins. Mol Cancer Ther 2005; 1: 61–69.
Gavert N, Vivanti A, Hazin J, Brabletz T, Ben-Ze’ev A . L1-Mediated colon cancer cell metastasis does not require changes in EMT and cancer stem cell markers. Mol Cancer Res 2011; 9: 14–24.
Vaid M, Prasad R, Sun Q, Katiyar SK . Silymarin targets β-catenin signaling in blocking migration/invasion of human melanoma cells. PLoS One 2011; 6: e23000.
Easwaran V, Lee SH, Inge L, Guo L, Goldbeck C, Garrett E et al. Beta-Catenin regulates vascular endothelial growth factor expression in colon cancer. Cancer Res 2003; 63: 3145–3153.
Mao W, Millar JC, Wang WH, Silverman SM, Liu Y, Wordinger RJ et al. Existence of the canonical Wnt signaling pathway in the human trabecular meshwork. Invest Ophthalmol Vis Sci 2011; 53: 7043–7051.
Gauger KJ, Shimono A, Crisi GM, Schneider SS . Loss of SFRP1 promotes ductal branching in the murine mammary gland. BMC Dev Biol 2012; 12: 25.
Sturgeon SR, Balasubramanian R, Schairer C, Muss HB, Ziegler RG, Arcaro KF . Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics 2012; 7: 1258–1267.
Wang X, Wang H, Bu R, Fei X, Zhao C, Song Y . Methylation and aberrant expression of the Wnt antagonist secreted Frizzled-related protein 1 in bladder cancer. Oncol Lett 2012; 4: 334–338.
Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM et al. DNA methylation inhibitors in cancer: Recent and future approaches. Biochimie 2012; 94: 2280–2296.
Li KK, Li F, Li QS, Yang K, Jin BDNA . Methylation as a target of epigenetic therapeutics in cancer. Anticancer Agents Med Chem 2013; 13: 242–247.
Arora P, Kim EO, Jung JK, Jang KL . Hepatitis C virus core protein dowregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett 2008; 261: 244–252.
Gong C, Tao G, Yang L, Liu J, Liu Q, Li W et al. Methylation of PARP-1 promoter involved in the regulation of nano-Sio2-induced decrease of PARP-1 mRNA expression. Toxicol Lett 2012; 209: 264–269.
Chiam K, Centenera MM, Butler LM, Tilley WD, Bianco-Miotto T . GSTP1 DNA methylation and expression status is indicative of 5-aza-2′-deoxycytidine efficacy in human prostate cancer cells. PLoS One 2011; 6: e25634.
Benegiamo G, Vinciguerra M, Mazzoccoli G, Piepoli A, Andriulli A, Pazienza V . DNA methyltransferases 1 and 3b expression in Huh-7 cells expressing HCV core protein of different genotypes. Dig Dis Sci 2012; 57: 1598–1603.
Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P . Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 1997; 275: 1790–1792.
Chan EF, Gat U, McNiff JM, Fuchs E . A common human skin tumour is caused by activating mutations in beta-catenin. Nat Genet 1999; 21: 410–413.
Campbell PM, Szyf M . Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway. Carcinogenesis 2003; 24: 17–24.
Kaneda A, Matsusaka K, Aburatani H, Fukayama M . Epstein-Barr virous infection as an epigenetic of tumorigenesis. Cancer Res 2012; 72: 3445–3450.
Milavetz B, Kallestad L, Gefroh A, Adams N, Woods E, Balakrishnan L . Virion-mediated transfer of SV40 epigenetic information. Epigenetics 2012; 7: 528–534.
Wu DW, Tsai LH, Chen PM, Lee MC, Wang L, Chen CY et al. Loss of TIMP-3 promotes tumor invasion via elevated IL-6 production and predicts poor survival and relapse in HPV-infected non-small cell lung cancer. Am J Pathol 2012; 181: 1796–1806.
Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS . Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med 2012; 13: 127–134.
Lee JY, Lee TH . Effects of DNA methylation on the structure of nucleosomes. J Am Chem Soc 2012; 134: 173–175.
Rountree MR, Bachman KE, Baylin SB . DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 2000; 25: 269–277.
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T . DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 2000; 24: 88–91.
Cervoni N, Szyf M . Demethylase activity is directed by histone acetylation. J Biol Chem 2001; 276: 40778–40787.
Akkari L, Grégoire D, Floc’h N, Moreau M, Hernandez C, Simonin Y et al. Hepatitis C viral protein NS5A induces EMT and participates in oncogenic transformation of primary hepatocyte precursors. J Hepatol 2012; 57: 1021–1028.
Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL et al. Liver cancer-derived hepatitis C virus core proteins shift TGF-beta responses from tumor suppression to epithelial-mesenchymal transition. PLoS One 2009; 4: e4355.
Bose SK, Meyer K, Di Bisceglie AM, Ray RB, Ray R . Hepatitis C virus induces epithelial mesenchymal transition in primary human hepatocytes. J Virol 2012; 86: 13621–13628.
Su HY, Lai HC, Lin YW, Liu CY, Chen CK, Chou YC et al. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer 2010; 127: 555–567.
Tang N, Song WX, Luo J, Luo X, Chen J, Sharff KA et al. BMP-9-induced osteogenic differentiation of mesenchymal progenitors requires functional canonical Wnt/beta-catenin signalling. J Cell Mol Med 2009; 13: 2448–2464.
Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005; 11: 791–796.
Acknowledgements
This study was supported by research grants from Major National S&T program (2013ZX10002002, AH; 2012ZX10002005-003-002, NT), China National Natural Science Foundation (#30972586, #31171307, NT; #81171563, ZZ) and Natural Science Foundation Project of CQMU (NT). We thank Dr T-C He of University of Chicago, USA, for kind provision of adenoviruses AdGFP, AdsiDnmt1 and AdSFRP1. We are also grateful for Dr Takaji Wakita (National Institute of Infectious Diseases, Tokyo) for providing the HCV JFH-1 cDNA clone.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Quan, H., Zhou, F., Nie, D. et al. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial–mesenchymal transition. Oncogene 33, 2826–2835 (2014). https://doi.org/10.1038/onc.2013.225
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.225
Keywords
This article is cited by
-
Hepatocellular Carcinoma in the Era of Direct Antiviral Agents Against Hepatitis C Virus
Current Hepatology Reports (2024)
-
Major genomic mutations driving hepatocellular carcinoma
Genome Instability & Disease (2023)
-
The interaction of canonical Wnt/β-catenin signaling with protein lysine acetylation
Cellular & Molecular Biology Letters (2022)
-
TCF3 Induces DNMT1 Expression to Regulate Wnt Signaling Pathway in Glioma
Neurotoxicity Research (2022)
-
HCV Core protein represses DKK3 expression via epigenetic silencing and activates the Wnt/β-catenin signaling pathway during the progression of HCC
Clinical and Translational Oncology (2022)