Abstract
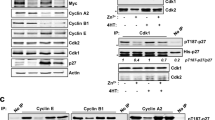
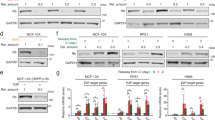
c-Myc is an oncogenic transcription factor capable of activating transcription by all three nuclear RNA polymerases, thus acting as a high-level coordinator of protein synthesis capacity and cell growth rate. c-Myc recruits RNA polymerase I-related transcription factors to the rDNA when quiescent cells are stimulated to re-enter the cell cycle. Using a model system of cell lines with variable c-Myc status, we show that on stimulation c-Myc rapidly induces gene loop structures in rDNA chromatin that juxtapose upstream and downstream rDNA sequences. c-Myc activation is both necessary and sufficient for this change in rDNA chromatin conformation. c-Myc activation induces association of TTF-1 with the rDNA, and c-Myc is physically associated with induced rDNA gene loops. The origins of two or more rDNA gene loops are closely juxtaposed, suggesting the possibility that c-Myc induces nucleolar chromatin hubs. Induction of rDNA gene loops may be an early step in the reprogramming of quiescent cells as they re-enter the growth cycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ansari A, Hampsey M . (2005). A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19: 2969–2978.
Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K et al. (2005). c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310.
Bukhari MH, Niazi S, Khan SA, Hashmi I, Perveen S, Qureshi SS et al. (2007). Modified method of AgNOR staining for tissue and interpretation in histopathology. Int J Exp Pathol 88: 47–53.
Cassidy BG, Yang-Yen HF, Rothblum LI . (1987). Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol 7: 2388–2396.
Comai L, Tanese N, Tjian R . (1992). The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68: 965–976.
de Laat W, Grosveld F . (2003). Spatial organization of gene expression: the active chromatin hub. Chromosome Res 11: 447–459.
Dean M, Levine RA, Ran W, Kindy MS, Sonenshein GE, Campisi J . (1986). Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem 261: 9161–9166.
Fischer AH, Bardarov Jr S, Jiang Z . (2004). Molecular aspects of diagnostic nucleolar and nuclear envelope changes in prostate cancer. J Cell Biochem 91: 170–184.
Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JC . (2005). TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J Biol Chem 280: 29551–29558.
Gomez-Roman N, Grandori C, Eisenman RN, White RJ . (2003). Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294.
Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J et al. (2006). Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp 73: 141–154.
Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN et al. (2005). c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318.
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA . (2005). Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302.
Grummt I . (2003). Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702.
Hernandez-Verdun D, Roussel P, Gebrane-Younes J . (2002). Emerging concepts of nucleolar assembly. J Cell Sci 115: 2265–2270.
Holzel M, Kohlhuber F, Schlosser I, Holzel D, Luscher B, Eick D . (2001). Myc/Max/Mad regulate the frequency but not the duration of productive cell cycles. EMBO Rep 2: 1125–1132.
Iritani BM, Eisenman RN . (1999). c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA 96: 13180–13185.
Keppel F . (1986). Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J Mol Biol 187: 15–21.
Kim S, Li Q, Dang CV, Lee LA . (2000). Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci USA 97: 11198–11202.
Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A et al. (2007). Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9: 45–56.
Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI . (2006). Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res 66: 4591–4601.
Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI . (1995). A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23: 1686–1690.
Lutz W, Leon J, Eilers M . (2002). Contributions of Myc to tumorigenesis. Biochim Biophys Acta 1602: 61–71.
Mais C, Wright JE, Prieto JL, Raggett SL, McStay B . (2005). UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64.
Mateyak MK, Obaya AJ, Adachi S, Sedivy JM . (1997). Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 8: 1039–1048.
Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G . (2008). Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J 27: 1255–1265.
O'Sullivan AC, Sullivan GJ, McStay B . (2002). UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22: 657–668.
O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J et al. (2004). Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018.
Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K et al. (2004). MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J 23: 3325–3335.
Ruggero D, Pandolfi PP . (2003). Does the ribosome translate cancer? Nat Rev Cancer 3: 179–192.
Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D . (2003). A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res 31: 6148–6156.
Sears RC . (2004). The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3: 1133–1137.
Seo J, Lozano MM, Dudley JP . (2005). Nuclear matrix binding regulates SATB1-mediated transcriptional repression. J Biol Chem 280: 24600–24609.
Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG et al. (2005). Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16: 2395–2413.
Splinter E, Grosveld F, de Laat W . (2004). 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol 375: 493–507.
Yin X, Giap C, Lazo JS, Prochownik EV . (2003). Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22: 6151–6159.
Acknowledgements
We thank Lars-Gunnar Larsson for the provision of rat fibroblasts HO15.15, TGR1 and Rat1MycER and Professor Dirk Eick for the generous gift of Smoxi4 cells. This work was supported by a Taiwan Fellowship from the Taiwan Root Medical Peace Corps, the Swedish Cancer Society and the research foundations of the Karolinska Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shiue, CN., Berkson, R. & Wright, A. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene 28, 1833–1842 (2009). https://doi.org/10.1038/onc.2009.21
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2009.21
Keywords
This article is cited by
-
Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation
Communications Biology (2019)
-
Nucleolus and chromatin
Histochemistry and Cell Biology (2018)
-
Chromatin loops and causality loops: the influence of RNA upon spatial nuclear architecture
Chromosoma (2017)
-
Nucleolar DNA: the host and the guests
Histochemistry and Cell Biology (2016)
-
Targeting RNA polymerase I to treat MYC-driven cancer
Oncogene (2015)