Abstract
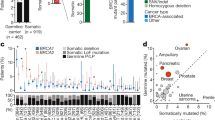
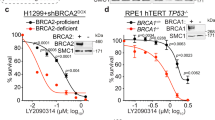
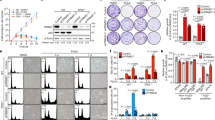
The BRCA1 and BRCA2 proteins are involved in the maintenance of genome stability and germ-line loss-of-function mutations in either BRCA1 or BRCA2 strongly predispose carriers to cancers of the breast and other organs. It has been demonstrated previously that inhibiting elements of the cellular DNA maintenance pathways represents a novel therapeutic approach to treating tumors in these individuals. Here, we show that inhibition of the telomere-associated protein, Tankyrase 1, is also selectively lethal with BRCA deficiency. We also demonstrate that the selectivity caused by inhibition of Tankyrase 1 is associated with an exacerbation of the centrosome amplification phenotype associated with BRCA deficiency. We propose that inhibition of Tankyrase 1 could be therapeutically exploited in BRCA-associated cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ame JC, Spenlehauer C, de Murcia G . (2004). The PARP superfamily. Bioessays 26: 882–893.
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E et al. (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917.
Chang W, Dynek JN, Smith S . (2003). TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev 17: 1328–1333.
Chang W, Dynek JN, Smith S . (2005). NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J 391: 177–184.
Chi NW, Lodish HF . (2000). Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem 275: 38437–38444.
d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T et al. (2003). A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198.
de Lange T . (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110.
DelloRusso C, Welcsh PL, Wang W, Garcia RL, King MC, Swisher EM . (2007). Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol Cancer Res 5: 35–45.
Dynek JN, Smith S . (2004). Resolution of sister telomere association is required for progression through mitosis. Science 304: 97–100.
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB et al. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921.
Iorns E, Lord CJ, Turner N, Ashworth A . (2007). Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov 6: 556–568.
Kaelin Jr WG . (2005). The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 5: 689–698.
Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW et al. (2008). Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32: 57–69.
McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S et al. (2006). Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66: 8109–8115.
Nigg EA . (2002). Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2: 815–825.
Seimiya H . (2006). The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer 94: 341–345.
Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T . (2005). Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell 7: 25–37.
Seimiya H, Muramatsu Y, Smith S, Tsuruo T . (2004). Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol Cell Biol 24: 1944–1955.
Seimiya H, Smith S . (2002). The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem 277: 14116–14126.
Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ et al. (2005). Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 37: 1281–1288.
Smith S, de Lange T . (1999). Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci 112 (Part 21): 3649–3656.
Smith S, Giriat I, Schmitt A, de Lange T . (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282: 1484–1487.
Turner N, Tutt A, Ashworth A . (2004). Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4: 814–819.
Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J et al. (1999). Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol 9: 1107–1110.
Tutt AN, Lord CJ, McCabe N, Farmer H, Turner N, Martin NM et al. (2005). Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol 70: 139–148.
Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW et al. (1999). Centrosome amplification and a defective G2–M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell 3: 389–395.
Acknowledgements
This work was funded by Breakthrough Breast Cancer and Cancer Research UK. We thank Jill Williamson and Dave Robertson for assistance with karyotyping and microscopy, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Rights and permissions
About this article
Cite this article
McCabe, N., Cerone, M., Ohishi, T. et al. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene 28, 1465–1470 (2009). https://doi.org/10.1038/onc.2008.483
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2008.483
Keywords
This article is cited by
-
PARP3, a new therapeutic target to alter Rictor/mTORC2 signaling and tumor progression in BRCA1-associated cancers
Cell Death & Differentiation (2019)
-
Identification and preclinical characterization of a novel and potent poly (ADP-ribose) polymerase (PARP) inhibitor ZYTP1
Cancer Chemotherapy and Pharmacology (2018)
-
Tankyrase inhibition impairs directional migration and invasion of lung cancer cells by affecting microtubule dynamics and polarity signals
BMC Biology (2016)
-
Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nature Biotechnology (2012)
-
Tankyrase-targeted therapeutics: expanding opportunities in the PARP family
Nature Reviews Drug Discovery (2012)