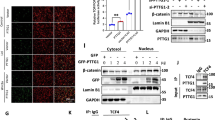
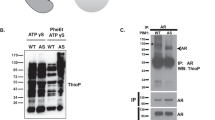
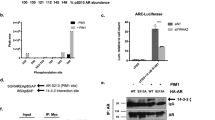
Abstract
Pituitary tumor transforming gene 1 (PTTG1), a transforming gene highly expressed in several cancers, is a mammalian securin protein regulating both G1/S and G2/M phases. Using protein array screening, we showed PTTG1 interacting with Aurora kinase A (Aurora-A), and confirmed the interaction using co-immunoprecipitation, His-tagged pull-down assays and intracellular immunofluorescence colocalization. PTTG1 transfection into HCT116 cells prevented Aurora-A T288 autophosphorylation, inhibited phosphorylation of the histone H3 Aurora-A substrate and resulted in abnormally condensed chromatin. PTTG1-null cell proliferation was more sensitive to Aurora-A knock down and to Aurora kinase Inhibitor III treatment. The results indicate that PTTG1 and Aurora-A interact to regulate cellular responses to anti-neoplastic drugs. PTTG1 knockdown is therefore a potential approach to improve the efficacy of tumor Aurora kinase inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Abbreviations
- Aurora-A:
-
Aurora kinase A
- PTTG1:
-
pituitary tumor transforming gene 1
References
Akino K, Akita S, Mizuguchi T, Takumi I, Yu R, Wang XY et al. (2005). A novel molecular marker of pituitary tumor transforming gene involves in a rat liver regeneration. J Surg Res 129: 142–146.
Bernal JA, Hernandez A . (2007). p53 stabilization can be uncoupled from its role in transcriptional activation by loss of PTTG1/securin. J Biochem (Tokyo) 141: 737–745.
Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F et al. (2002). Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet 32: 306–311.
Bernal JA, Roche M, Mendez-Vidal C, Espina A, Tortolero M, Pintor-Toro JA . (2008). Proliferative potential after DNA damage and non-homologous end joining are affected by loss of securin. Cell Death Differ 15: 202–212.
Boelaert K, Tannahill LA, Bulmer JN, Kachilele S, Chan SY, Kim D et al. (2003). A potential role for PTTG/securin in the developing human fetal brain. FASEB J 17: 1631–1639.
Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates III JR et al. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172.
Chen SS, Chang PC, Cheng YW, Tang FM, Lin YS . (2002). Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function. EMBO J 21: 4491–4499.
Chien W, Pei L . (2000). A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem 275: 19422–19427.
Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H et al. (2002). Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol 22: 874–885.
Donangelo I, Gutman S, Horvath E, Kovacs K, Wawrowsky K, Mount M et al. (2006). Pituitary tumor transforming gene overexpression facilitates pituitary tumor development. Endocrinology 147: 4781–4791.
El-Naggar SM, Malik MT, Kakar SS . (2007). Small interfering RNA against PTTG: a novel therapy for ovarian cancer. Int J Oncol 31: 137–143.
Eyers PA, Erikson E, Chen LG, Maller JL . (2003a). A novel mechanism for activation of the protein kinase Aurora A. Curr Biol 13: 691–697.
Eyers PA, Erikson E, Chen LG, Maller JL . (2003b). A novel mechanism for activation of the protein kinase Aurora A. Curr Biol 13: 691–697.
Gadea BB, Ruderman JV . (2005). Aurora kinase inhibitor ZM447439 blocks chromosome-induced spindle assembly, the completion of chromosome condensation, and the establishment of the spindle integrity checkpoint in Xenopus egg extracts. Mol Biol Cell 16: 1305–1318.
Gil-Bernabe AM, Romero F, Limon-Mortes MC, Tortolero M . (2006). Protein phosphatase 2A stabilizes human securin, whose phosphorylated forms are degraded via the SCF ubiquitin ligase. Mol Cell Biol 26: 4017–4027.
Heaney AP, Horwitz GA, Wang Z, Singson R, Melmed S . (1999). Early involvement of estrogen-induced pituitary tumor transforming gene and fibroblast growth factor expression in prolactinoma pathogenesis. Nat Med 5: 1317–1321.
Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S . (2000). Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet 355: 716–719.
Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M et al. (2003). Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114: 585–598.
Horn V, Thelu J, Garcia A, biges-Rizo C, Block MR, Viallet J . (2007). Functional interaction of Aurora-A and PP2A during mitosis. Mol Biol Cell 18: 1233–1241.
Hornig NC, Knowles PP, McDonald NQ, Uhlmann F . (2002). The dual mechanism of separase regulation by securin. Curr Biol 12: 973–982.
Hutterer A, Berdnik D, Wirtz-Peitz F, Zigman M, Schleiffer A, Knoblich JA . (2006). Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell 11: 147–157.
Ikezoe T, Yang J, Nishioka C, Tasaka T, Taniguchi A, Kuwayama Y et al. (2007). A novel treatment strategy targeting Aurora kinases in acute myelogenous leukemia. Mol Cancer Ther 6: 1851–1857.
Kakar SS, Malik MT . (2006). Suppression of lung cancer with siRNA targeting PTTG. Int J Oncol 29: 387–395.
Kim DS, Buchanan MA, Stratford AL, Watkinson JC, Eggo MC, Franklyn JA et al. (2006). PTTG promotes a novel VEGF–KDR–ID3 autocrine mitogenic pathway in thyroid cancer. Clin Otolaryngol 31: 246.
Kim DS, Franklyn JA, Smith VE, Stratford AL, Pemberton HN, Warfield A et al. (2007). Securin induces genetic instability in colorectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis 28: 749–759.
Krystyniak A, Garcia-Echeverria C, Prigent C, Ferrari S . (2006). Inhibition of Aurora A in response to DNA damage. Oncogene 25: 338–348.
Lai Y, Xin D, Bai J, Mao Z, Na Y . (2007). The important anti-apoptotic role and its regulation mechanism of PTTG1 in UV-induced apoptosis. J Biochem Mol Biol 40: 966–972.
Li JJ, Li SA . (2006). Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol Ther 111: 974–984.
Li M, York JP, Zhang P . (2007). Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol Cell Biol 27: 3481–3488.
Lindon C, Pines J . (2004). Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol 164: 233–241.
Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y et al. (2002). Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells 7: 1173–1182.
McCabe CJ, Khaira JS, Boelaert K, Heaney AP, Tannahill LA, Hussain S et al. (2003). Expression of pituitary tumour transforming gene (PTTG) and fibroblast growth factor-2 (FGF-2) in human pituitary adenomas: relationships to clinical tumour behaviour. Clin Endocrinol (Oxf) 58: 141–150.
Mountzios G, Terpos E, Dimopoulos MA . (2008). Aurora kinases as targets for cancer therapy. Cancer Treat Rev 34: 175–182.
Ohashi S, Sakashita G, Ban R, Nagasawa M, Matsuzaki H, Murata Y et al. (2006). Phospho-regulation of human protein kinase Aurora-A: analysis using anti-phospho-Thr288 monoclonal antibodies. Oncogene 25: 7691–7702.
Pascreau G, Arlot-Bonnemains Y, Prigent C . (2003). Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res 5: 369–374.
Pei L . (1999). Pituitary tumor-transforming gene protein associates with ribosomal protein S10 and a novel human homologue of DnaJ in testicular cells. J Biol Chem 274: 3151–3158.
Pei L . (2000). Activation of mitogen-activated protein kinase cascade regulates pituitary tumor-transforming gene transactivation function. J Biol Chem 275: 31191–31198.
Pei L . (2001). Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. J Biol Chem 276: 8484–8491.
Pei L, Melmed S . (1997). Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol 11: 433–441.
Pereira G, Schiebel E . (2003). Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302: 2120–2124.
Prigent C, Dimitrov S . (2003). Phosphorylation of serine 10 in histone H3, what for? J Cell Sci 116: 3677–3685.
Saez C, Japon MA, Ramos-Morales F, Romero F, Segura DI, Tortolero M et al. (1999). hpttg is over-expressed in pituitary adenomas and other primary epithelial neoplasias. Oncogene 18: 5473–5476.
Sheleg SV, Peloponese JM, Chi YH, Li Y, Eckhaus M, Jeang KT . (2007). Evidence for co-operative transforming activity of human pituitary tumor transforming gene (PTTG) and HTLV-1 Tax. J Virol 81: 7894–7901.
Solbach C, Roller M, Fellbaum C, Nicoletti M, Kaufmann M . (2004). PTTG mRNA expression in primary breast cancer: a prognostic marker for lymph node invasion and tumor recurrence. Breast 13: 80–81.
Sun C, Chan F, Briassouli P, Linardopoulos S . (2007). Aurora kinase inhibition downregulates NF-kappaB and sensitises tumour cells to chemotherapeutic agents. Biochem Biophys Res Commun 352: 220–225.
Tarabykin V, Britanova O, Fradkov A, Voss A, Katz LS, Lukyanov S et al. (2000). Expression of PTTG and prc1 genes during telencephalic neurogenesis. Mech Dev 92: 301–304.
Tong Y, Tan Y, Zhou C, Melmed S . (2007). Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene 26: 5596–5605.
Vlotides G, Eigler T, Melmed S . (2007). Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev 28: 165–186.
Vogel C, Hager C, Bastians H . (2007). Mechanisms of mitotic cell death induced by chemotherapy-mediated G2 checkpoint abrogation. Cancer Res 67: 339–345.
Vogel C, Kienitz A, Muller R, Bastians H . (2005). The mitotic spindle checkpoint is a critical determinant for topoisomerase-based chemotherapy. J Biol Chem 280: 4025–4028.
Waizenegger I, Gimenez-Abian JF, Wernic D, Peters JM . (2002). Regulation of human separase by securin binding and autocleavage. Curr Biol 12: 1368–1378.
Walter AO, Seghezzi W, Korver W, Sheung J, Lees E . (2000). The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 19: 4906–4916.
Wang H, Liu D, Wang Y, Qin J, Elledge SJ . (2001a). Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes Dev 15: 1361–1372.
Wang Z, Melmed S . (2000). Pituitary tumor transforming gene (PTTG) transforming and transactivation activity. J Biol Chem 275: 7459–7461.
Wang Z, Moro E, Kovacs K, Yu R, Melmed S . (2003). Pituitary tumor transforming gene-null male mice exhibit impaired pancreatic beta cell proliferation and diabetes. Proc Natl Acad Sci USA 100: 3428–3432.
Wang Z, Yu R, Melmed S . (2001b). Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol Endocrinol 15: 1870–1879.
Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X . (2008). GPS 2.0: prediction of kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics; e-pub ahead of print.
Yang H, He L, Kruk P, Nicosia SV, Cheng JQ . (2006). Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer 119: 2304–2312.
Yu R, Lu W, Chen J, McCabe CJ, Melmed S . (2003). Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology 144: 4991–4998.
Zhang D, Hirota T, Marumoto T, Shimizu M, Kunitoku N, Sasayama T et al. (2004). Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene 23: 8720–8730.
Zhang D, Shimizu T, Araki N, Hirota T, Yoshie M, Ogawa K et al. (2008). Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice. Oncogene; e-pub ahead of print.
Zhang Y, Ni J, Huang Q, Ren W, Yu L, Zhao S . (2007). Identification of the auto-inhibitory domains of Aurora-A kinase. Biochem Biophys Res Commun 357: 347–352.
Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A et al. (1998). Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet 20: 189–193.
Zou H, McGarry TJ, Bernal T, Kirschner MW . (1999). Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285: 418–422.
Acknowledgements
PTTG1 WT and KO HCT116 cells were kindly provided by Dr Bert Vogelstein, Johns Hopkins University. PTTG1-GFP and PTTG1-PCDNA3.1 plasmids were kindly provided by Dr Run Yu. Supported by NIH Grant CA 75979 (SM), T32 DK007770, and The Doris Factor Molecular Endocrinology Laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure
The authors state no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary information
Rights and permissions
About this article
Cite this article
Tong, Y., Ben-Shlomo, A., Zhou, C. et al. Pituitary tumor transforming gene 1 regulates Aurora kinase A activity. Oncogene 27, 6385–6395 (2008). https://doi.org/10.1038/onc.2008.234
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2008.234
Keywords
This article is cited by
-
LRRK2 Pathways Leading to Neurodegeneration
Current Neurology and Neuroscience Reports (2015)
-
Altered expression of securin (Pttg1) and serpina3n in the auditory system of hearing-impaired Tff3-deficient mice
Cellular and Molecular Life Sciences (2011)